Adenylyl cyclases
| Adenylyl cyclases | ||
|---|---|---|

|
||
| Schematic representation of adenylyl cyclase, embedded in the cell membrane | ||
| Identifier | ||
| Gene name (s) | ADCY1 , ADCY2 , ADCY3 , ADCY4 , ADCY5 , ADCY6 , ADCY7 , ADCY8 , ADCY9 , ADCY10 | |
| Enzyme classification | ||
| EC, category | 4.6.1.1 , lyase | |
| Substrate | ATP | |
| Products | cAMP + diphosphate | |
| Occurrence | ||
| Parent taxon | Creature | |
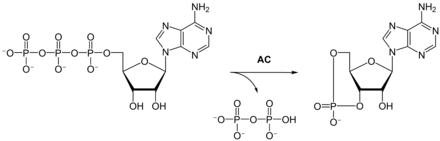
Adenylyl cyclases ( AC ), formerly adenylate cyclases , belong to the class of lyases , i. H. molecule-splitting enzymes .
In higher organisms they are important mediators between hormones or other messenger substances that bind to the outside of the cell membrane , and cell-internal messenger substances ( second messenger , "secondary messenger substance"), which transmit the effect of these hormones within the cell.
In the process of signal transduction , the hormone first binds to a suitable G protein-coupled receptor . This releases the corresponding G-protein , which in turn activates adenylyl cyclase. Inside the cell, this then forms the secondary messenger substance cyclic adenosine monophosphate (cAMP) from adenosine triphosphate (ATP).
Adenylyl cyclases occur in almost all living things. In humans, ten isoenzymes are currently known, mostly bound to the cell membrane .
Occurrence
Adenylyl cyclases are widespread in nature. Even simple unicellular eukaryotic organisms such as Paramecium , Euglena and Dictyostelium as well as some prokaryotes use adenylyl cyclases to convert ATP into cAMP. In addition, some prokaryotic pathogens use the function of adenylyl cyclases or the cAMPs of higher living organisms by forming and releasing adenylyl cyclases or activators of adenylyl cyclases themselves. For example, the invasiveness of the cholera pathogen Vibrio cholerae is attributed to cholera toxin , which indirectly activates adenylyl cyclases . The released adenylyl cyclases from Bordetella pertussis and Bacillus anthracis , which are only activated in the body of the host organism, are part of their toxic principle.
Classification
The adenylyl cyclases belong to the group of lyases. Classically, the adenylyl cyclases are divided into three main classes. Class I includes the adenylyl cyclases of gram-negative bacteria. Class II adenylyl cyclases play a role as toxic proteins of prokaryotic pathogens and are dependent on the host organism's calmodulin protein . The adenylyl cyclases of class III are numerically largest. It includes all eukaryotic adenylyl cyclases as well as numerous prokaryotic enzymes. Another three classes (IV-VI) of adenylyl cyclases are mentioned less frequently.
The human adenylyl cyclases comprise 10 different isoenzymes and are referred to as AC 1 to AC 10 (or AC I to AC X). The isoenzymes AC 1 to AC 9 are membrane-bound proteins. The isoenzyme AC 10 , also known as soluble adenylyl cyclase (sAC, from soluble adenylyl cyclase ), is a cytosolic protein that occurs predominantly in special cell compartments. In addition, the isoenzymes of the adenylyl cyclases differ in their regulatory mechanisms.
| AC 1 | AC 2 | AC 3 | AC 4 | AC 5 | AC 6 | AC 7 | AC 8 | AC 9 | AC 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| synonym | sAC | |||||||||
| Occurrence | Brain, adrenal glands | Brain, lungs, skeletal muscles | Olfactory epithelium, brain, adrenal glands, adipose tissue, pancreas | ubiquitous | Heart, brain | Heart, kidneys, brain, liver | Brain, platelets, heart, spleen, lungs | Brain, lungs | Skeletal muscles, heart, brain, pancreas | Germ cells |
| genetics | ||||||||||
| Gene name | ADCY1 | ADCY2 | ADCY3 | ADCY4 | ADCY5 | ADCY6 | ADCY7 | ADCY8 | ADCY9 | ADCY10 |
| Gene locus | 7p13-p12 | 5p15.3 | 2p24-p22 | 14q11.2 | 3q13.2-q21 | 12q12-q13 | 16q12.1 | 8q24 | 16p13.3 | 1q24 |
| protein | ||||||||||
| UniProt-Bez. | Q08828 | Q08462 | O60266 | Q8NFM4 | O95622 | O43306 | P51828 | P40145 | O60503 | Q96PN6 |
| Length (amino acids) | 1119 | 1091 | 1144 | 1077 | 1261 | 1168 | 1080 | 1251 | 1353 | 1610 |
| Regulation * | ||||||||||
| Gα s | + | + | + | + | + | + | + | + | + | O |
| Gα i / o | - | - | - | - | - | O | ||||
| Gβγ | - | + | O | + | + | - | O | |||
| Ca 2+ / ( calmodulin ) | + | O | + | O | - | - | O | + | + | |
| Adenosine | - | - | - | - | - | - | - | - | - | - |
| Forskolin | + | + | + | + | + | + | + | + | O | O |
* + Activation, o without effect, - inhibition; Ca 2+ effects at low (<1 µM) Ca 2+ concentrations
structure
The adenylyl cyclases occurring in living nature sometimes follow very different construction plans. What they all have in common is that they have at least one cyclase homology domain that is essential for their function . The membrane-bound adenylyl cyclases of multicellular animals ( Metazoa ) consist of two transmembrane domains each with six α-helices (M 1 and M 2 ) and two cytosolic cyclase homology domains (C 1 and C 2 ). The two approximately 40 kDa cyclase homology domains can be further subdivided into catalytic (C 1a and C 2a ) and regulatory subdomains (C 1b and C 2b ). The catalytic subdomains show a high degree of homology .
The three-dimensional structure of the catalytic subdomains of different animal adenylyl cyclases could be elucidated with the help of the X-ray structure analysis. With the help of this technique, a structure consisting of a βαββαβ motif, which, for example, also occurs in some DNA polymerases , could be confirmed for the catalytic subdomains . Both catalytic domains interact with each other to form a functional unit. On the contact side of the two subunits, the C 1a -C 2a unit formed has a groove with a binding site for the substrate ATP. This furrow also contains another binding site for the activator forskolin , a diterpene from the harp bush Plectranthus barbatus . The binding site for the stimulating G proteins G s and G olf comprises a small portion of the N terminus of the C 1a subdomain and a larger portion of the C 2a subdomain. In addition, another binding site on the regulatory C 1b subdomain is suspected. The postulated binding site for inhibitory G proteins (G i / o family) on the C 1a subdomain is different from the binding site for stimulating G proteins.
function
Catalysis of cAMP production
Adenylyl cyclase catalyzes the formation of cAMP from ATP with elimination of pyrophosphate . Depending on the isoenzyme, one molecule of adenylyl cyclase converts between one and 100 substrate molecules per second. The respective Michaelis constants K m , which correspond to the substrate concentration at half the maximum conversion rate, are between 30 and 400 μM.
In vertebrates and invertebrates (e.g. Aplysia ), cAMP functions as a second messenger . The enzyme is therefore part of the signal cascade for the transmission of signals within cells . It is also often associated with stimulating transmitter release and signal conduction, increasing heart rate, and smooth muscle relaxation. The adenylyl cyclase is therefore responsible for the transmission of stimuli and for the effects of numerous hormones ( e.g. glucagon and adrenaline ) and neurotransmitters (e.g. serotonin ).
regulation
In the human organism, the activity of the adenylyl cyclases is physiologically regulated, in particular by the G proteins involved in signal transduction. Stimulating G proteins of the G s / olf family favor a closed conformation of the dimeric catalytic unit of adenylyl cyclase and thus activate its enzyme function. Inhibitory G i / o proteins lead to an inhibition of the adenylyl cyclase function via a stabilization of the open conformation. Some isoenzymes of adenylyl cyclase (AC 1 , AC 3 and AC 8 ) are also regulated by calcium / calmodulin .
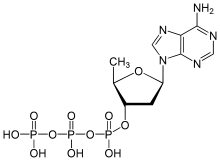
In addition, the adenylyl cyclases can be regulated pharmacologically. The diterpene forskolin is a non-selective activator of the adenylyl cyclases, which activates all human adenylyl cyclases with the exception of AC 9 . The discovery that adenosine nonselectively inhibits all adenylyl cyclases led to the development of numerous nucleoside and nucleotide analogues. As so-called P-site inhibitors of adenylyl cyclases , they usually lead to non-competitive or uncompetitive inhibition based on various mechanisms. For their inhibitory effect, the nucleoside analogs, such as, for example, 2'-deoxyadenosine , require the cell's own pyrophosphate as a co-inhibitor. Their inhibitory potency is exceeded by the pyrophosphate-independent nucleotide analogs such as 2 ', 5'-dideoxyadenosine-3'-tetraphosphate , 2', 5'-dideoxyadenosine-3'-triphosphate . In addition, the adenylyl cyclases can be competitively inhibited by nucleotide substrate analogs, such as, for example, β-L-2 ', 3'-dideoxy-5'-adenosine triphosphate , by displacing the substrate ATP.
Individual evidence
- ↑ Entry on adenylate cyclase. In: Römpp Online . Georg Thieme Verlag, accessed on June 13, 2011.
- ^ Cooper DM: Regulation and organization of adenylyl cyclases and cAMP . In: Biochem. J. . 375, No. Pt 3, November 2003, pp. 517-29. doi : 10.1042 / BJ20031061 . PMID 12940771 . PMC 1223734 (free full text).
- ↑ a b Roger A. Johnson: Adenylate cyclases . In: Walter Rosenthal; Offermanns, Stefan (Ed.): Encyclopedia of molecular pharmacology . Springer, Berlin 2008, ISBN 3-540-38916-4 , pp. 28-37.
- ↑ Zippin JH, Chen Y, Nahirney P, et al. : Compartmentalization of bicarbonate-sensitive adenylyl cyclase in distinct signaling microdomains . In: FASEB J . . 17, No. 1, January 2003, pp. 82-4. doi : 10.1096 / fj.02-0598fje . PMID 12475901 .
- ↑ a b c Tang WJ, Hurley JH: Catalytic mechanism and regulation of mammalian adenylyl cyclases . In: Mol. Pharmacol. . 54, No. 2, August 1998, pp. 231-40. PMID 9687563 .
- ^ Hurley JH: Structure, mechanism, and regulation of mammalian adenylyl cyclase . In: J. Biol. Chem. . 274, No. 12, March 1999, pp. 7599-7602. PMID 10075642 .
- ↑ Eric Kandel: Principles of Neural Science, Fourth Edition . McGraw-Hill Companies, Incorporated, 2000, ISBN 978-0-838-57701-1 ( limited preview in Google Book Search).
- ↑ Dessauer CW, Tesmer JJ, Sprang SR, Gilman AG: The interactions of adenylate cyclases with P-site inhibitors . In: Trends Pharmacol. Sci. . 20, No. 5, May 1999, pp. 205-10. PMID 10354616 .