Purine Biosynthesis
| Parent |
| Synthesis of IMP |
| Gene Ontology |
|---|
| QuickGO |
The purine biosynthesis (more precisely: IMP de novo synthesis ) is the metabolic pathway that all living beings capable of purines produce from simple starting materials. Purines are of paramount importance as a component of genetic information , as an energy carrier ( ATP ) and as the basis for the synthesis of other important substances.
The purines are not synthesized in the organism as free molecules , but always as nucleotides . The starting point of the purine synthesis is the α- D- ribose-5-phosphate , an intermediate product of the pentose phosphate cycle . The end product after eleven steps is inosine monophosphate (IMP), the nucleotide of hypoxanthine , which is converted into the nucleotides of xanthinosine , adenosine or guanosine in further steps .
In humans, purine synthesis takes place in all cell types that have an active cell nucleus .
Overview
The basic structure of the purine is built up step-by-step, with different molecules supplying the individual components:
Genomic organization in different species
The process and the genes of de novo synthesis are highly conserved . Although de novo synthesis is almost ubiquitous , there are differences between the species in the organization of the enzymes and the genes coding for them . The number of genes involved in the ten enzymatic steps decreases from prokaryotes to eukaryotes , but the complexity of the enzymes increases in the same way. So-called cluster genes are formed , which code for multifunctional enzymes .
In humans, the catalytic domains of the individual steps (2,5,3), (6,7) and (9,10) can each be found on a protein (PRPP synthesis counts as step zero).
Individual steps and intermediate products
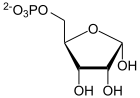
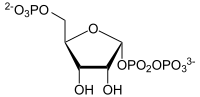
PRPP
α- D- ribose-5-phosphate is converted to α- D -5-phosphoribosyl-1-pyrophosphate (PRPP) by means of ribose phosphate diphosphokinase .
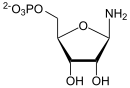
PRE
From PRPP and glutamine , 5- phosphoribosylamine (PRA) and glutamate are formed with the help of amidophosphoribosyltransferase . This reaction step is definitive, that is, from the products of this and the next reaction, only IMP can be produced from now on.
The reaction in equilibrium is shifted strongly to the right by hydrolysis of the diphosphate . IMP , GMP and AMP act as inhibitors .
AT ALL
The phosphoribosylamine glycine ligase - domain of the trifunctional purine synthesis protein facilitates the addition of glycine to 5-phosphoribosylamine (PRA) to form glycine amide ribonucleotide (GAR).
FGAR
The phosphoribosylglycine amide formyltransferase domain (GART) of the trifunctional purine synthesis protein catalyzes the formylation of glycine amide ribonucleotide (GAR) to formylglycine amide ribonucleotide (FGAR) using formyl tetrahydrofolate .
FGAM
Phosphoribosylformylglycine amide (FGAR) and glutamine are converted to phosphoribosylformylglycine amidine (FGAM) and glutamic acid. The catalyzing enzyme is FGAM synthase .
AIR
The phosphoribosylformylglycine amidine cyclo-ligase domain of the trifunctional purine synthesis protein cyclizes formylglycine amidine ribonucleotide (FGAM) to 5-aminoimidazole ribonucleotide (AIR).
CAIR
5-Aminoimidazole ribonucleotide (AIR) is carboxylated by ADE2 to 5-aminoimidazole-4-carboxylatribonucleotide (CAIR) using the phosphoribosylaminoimidazole carboxylase domain (AIRC) .
SAICAR
CAIR and aspartic acid are ligated, with consumption of ATP catalyzed by the SAICAR synthase domain of ADE2 .
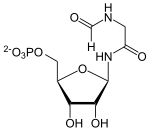
AICAR
Fumarate is split off from SAICAR, AICAR is formed by means of adenylosuccinate lyase .
FAICAR
AICAR is formylated to FAICAR. The catalyst is the AICAR formyltransferase domain of the bifunctional purine synthesis protein .
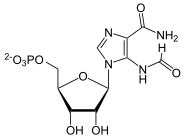
IMP
FAICAR cyclizes with elimination of water to IMP by means of the IMP cyclohydrolase domain of the bifunctional purine synthesis protein.
pathology
Several mutations are known in humans in genes that code for enzymes involved in purine synthesis and can lead to rare hereditary metabolic diseases :
Mutations in the PRPS1 gene of ribose phosphate diphosphokinase can lead to overactivity of the enzyme, which leads to an increased hereditary risk of gout . Other PRPS1 mutations reduce the enzyme activity and are the cause of the so-called Rosenberg-Chutorian syndrome and a form of deafness (ARTS).
Mutations in the ADSL gene of adenylosuccinate lyase are responsible for adenylosuccinate lyase deficiency , a rare hereditary disease.
Mutations in the ATIC gene of the bifunctional purine synthesis protein can cause the very rare severe AICA ribosiduria .
Individual evidence
- ^ Wagner, KG & Backer, AI (1992). Dynamics of nucleotides in plants studied on a cellular basis . Int. Rev. Cytol . 134, 184
- ↑ Jassal / reactome: 5-phospho-alpha-D-ribose 1-diphosphate (PRPP) + H2O + L-glutamine <=> 5-phosphoribosylamine + L-glutamate + pyrophosphate ( Memento of the original from May 13, 2016 in the Internet Archive ) Info: The archive link was inserted automatically and has not yet been checked. Please check the original and archive link according to the instructions and then remove this notice.
- ↑ UniProt P60891
- ↑ orphaNet: Ataxia - hearing loss - optic atrophy, fatal
- ↑ orphaNet: Adenylosuccinate lyase deficiency
- ↑ orphaNet: AICA ribosiduria