Cytochrome c oxidase
| Cytochrome c oxidase | ||
|---|---|---|
|
||
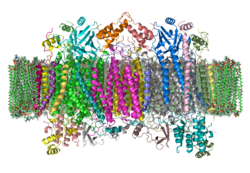
| Ribbon model of a bovine cytochrome c oxidase dimer in the membrane according to PDB 1OCC | ||
| Transporter classification | ||
| TCDB | 3.D.4.7.1 | |
| designation | proton transferring COX | |
| Enzyme classification | ||
| EC, category | 1.9.3.1 , oxidoreductase | |
| Response type | Redox reaction | |
| Substrate | 4 cytochrome c (reduced) + O 2 + 8 H + (in) | |
| Products | 4 cytochrome c (oxidized) + 2 H 2 O + 4 H + (out) | |
| Occurrence | ||
| Parent taxon | Creature | |
| Parent |
| Respiratory chain complex |
| Gene Ontology |
|---|
| QuickGO |
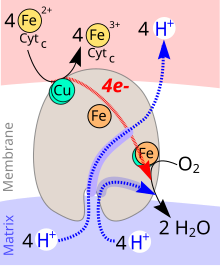
The enzyme cytochrome c oxidase (COX) , more precisely cytochrome c : oxygen oxidoreductase (systematic name), cytochrome aa 3 complex or complex IV of the mitochondrial respiratory chain , is an oxidoreductase . The enzyme complex , which consists of three subunits in bacteria and thirteen subunits in eukaryotes, catalyzes in a coupled reaction the oxidation of cytochrome c with the reduction of oxygen to water and the transport of protons across a biological membrane .
Mutations in the genes that code for the individual subunits ( MT-CO1 , MT-CO2 , MT-CO3 ) can cause rare hereditary diseases that are grouped under cytochrome c oxidase deficiency (MT-C4D), namely liver optic neuropathy (LHON), recurrent myoglobinuria , and mitochondrial nonsyndromal sensorineural hearing loss .
The cytochrome c oxidase belongs to the superfamily of heme-copper oxidases , which are the terminal electron acceptor of the respiratory chain in all aerobically breathing organisms . They are responsible for almost all of the oxygen consumption of the breathing organisms. The oxidases are stored in the inner mitochondrial membrane in eukaryotes and in the inner cell membrane in prokaryotes . Variants of cytochrome c oxidase occur in the cell membrane of aerobic bacteria . Some of these contain modified cofactors (heme variants) or use other electron donors than cytochrome c ( quinol oxidases, e.g. in Escherichia coli ). They all have great structural and functional homology and contain a heme group and a copper ion in the active center .
Catalyzed transport
The transport equation is:
4 Cyt c (Fe 2+ ) + O 2 + 8 H + inside → 4 Cyt c (Fe 3+ ) + 2 H 2 O + 4 H + outside
The function of cytochrome c oxidase consists of the
- Reduction of oxygen to water (biological oxyhydrogen reaction ) by means of electrons from cytochrome c and dem
- Transport of protons ( proton pump ) across the biological membrane .
During the catalytic cycle of cytochrome c oxidase, one molecule of oxygen (O 2 ) is reduced to two molecules of water (H 2 O). Four electrons (e - ) from four molecules of cytochrome c and protons (H + ) are used as reducing agents for water formation from the interior of the mitochondrion (matrix). The energy released during the reduction of oxygen to water is used to build up a proton gradient across the inner mitochondrial membrane. Four protons are transported from the interior of the mitochondrion into the intermembrane space per reaction cycle.
Recently, some ideas could be developed about the complex interplay of the oxygen chemistry taking place, the electron transfer reactions , as well as the proton uptake and pumping steps and their exact chronological sequences. In Fig. 3 and 4 there are models of the sequence of the redox reactions and the mechanics of the pumping mechanism.
Overall cycle in the reduction of O 2 to 2 H 2 O.
The cytochrome c oxidase is a membrane protein. It contains different metallic prosthetic groups :
- 2 heme groups
- The cytochromes a and a 3
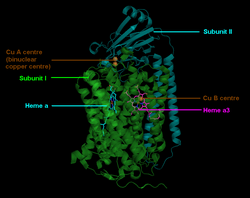
- Two copper centers (Cu A and Cu B ).
The Cu A is located at the binding site for cytochrome c . After its oxidation, the electron is passed on via cytochrome a to cytochrome a 3 , where it, together with Cu B, catalyzes the O 2 reduction. The process is shown in detail in Fig. 3.
- A :An O 2 moleculeis coordinatively bound to theFe (II) of heme a3 .
- → Two rearrangements begin (thin black dashed arrows).
- P M : Fe (II) isoxidizedto Fe (IV) = O 2− and Cu (I) to Cu (II) -OH - . The hydrogen atom comes from a tyrosine residue, which becomes a radical (*).
- → An e - attaches to the radical.
- P R : The radical is reduced to the tyrosine anion.
- → The first H + is translocated from inside to outside (blue dashed arrow). Cu (II) -OH - is protonated at the same time .
- Q : During the protonation, Cu (II) with coordinatively bound water was formed.
- → The second H + is translocated. Fe (IV) = O 2− is reduced and protonated. When Cu (II) is split off, the first water molecule is released.
- O H :Fe (III) -OH - is formedby protonation and reduction of Fe (IV) = O 2− .
- → Third proton transloction. Reduction of Cu (II) and protonation of the tyrosine anion.
- E H : Cu (II) is reduced to Cu (I) and the tyrosine anion is protonated to Tyr-OH.
- → Fourth H + translocation. Fe (III) -OH - is reduced and the OH - group is protonated.
- R : The central atom of heme a3 is reduced to Fe (II). The second H 2 O was created.
- → The two resulting water molecules are excreted. An oxygen atom is coordinatively attached to Fe (II).
Mechanism of proton translocation
The enzyme makes an important contribution to energy metabolism not only in mitochondria, but also in a large number of aerobic prokaryotes by acting as a proton pump to build up a chemiosmotic potential .
When four cytochrome c molecules are oxidized , four protons are translocated. Each of these reactions (to put it simply) follows the same mechanism. It is shown in four phases (1–4) on the right in Fig. 4.
- 1: The cytochrome c oxidase is embedded in the cell membrane (white). On the outside (reddish), reduced Cyt c accumulates on a copper center. The entrances to two proton channels D and K are located on the cell plasma side (bottom) of the membrane .
- 2: After an electron has been transferred from Cyt c to the Cu center, the electron is passed on to the Fe center of Cyt a . The D-channel is opened and a proton penetrates through it up to the “proton loading point” (PLS).
- 3: The electron moves from the cyt a to the cyt a3 center, while a proton penetrates the K channel. The PLS proton at the top of the enzyme experiences a significant increase in its acidity (from pK = 11 to pK = 5!).
- 4: The PLS proton is released on the outside of the membrane. Synchronously with this, the H + in the K channel undergoes one of the protonation reactions shown on the left, while the e - is consumed by the cyt a3 in one of the four reductions shown in Fig. 3.
structure
The mitochondrial enzyme complex IV in mammals consists of 13 subunits, of which the subunits I – III are mitochondrial and the other subunits IV – XIII are encoded by the nucleus. The subunit I has the three redox-active metal centers heme a , heme a 3 and Cu B . Heme a 3 and Cu B together form the catalytically active center, to which oxygen is bound and reduced to water. Subunit II has the redox-active metal center Cu A , which accepts electrons from cytochrome c , which are then transferred to heme a and on to heme a 3 .
Complex IV in humans in detail:
| number | Gene name | UniProt | Size (aa) |
OMIM | comment |
|---|---|---|---|---|---|
| 1 | MT-CO1 | P00395 | 513 | 516030 | Catalytic subunit; Heme , Cu 2+ ; Membrane domains; pathological mutations |
| 2 | MT-CO2 | P00403 | 227 | 516040 | Cu 2+ ; Membrane domains; pathological mutations |
| 1 | MT-CO3 | P00414 | 261 | 516050 | Membrane domains; pathological mutations |
| 1 | COX4I1 | P13073 | 169 | 123864 | |
| 1 | COX5A | P20674 | 150 | 603773 | Heme A |
| 1 | COX5B | P10606 | 98 | 123866 | Zn 2+ |
| 1 | COX6A1 | P12074 | 85 | 602072 | |
| 1 | COX6B1 | P14854 | 85 | 124089 | Pathological mutations |
| 1 | COX6C1 | P09669 | 74 | 124090 | Membrane domain |
| 1 | COX7A2L | O14548 | 59 | 605771 | |
| 1 | COX7B | P24311 | 56 | 603792 | Membrane domain |
| 1 | COX7C | P15954 | 47 | 603774 | Membrane domain |
| 1 | COX8A | P10176 | 44 | 123870 | Membrane domain |
Inhibitors
Cyanides , carbon monoxide , hydrogen sulfide and azides are inhibitors of cytochrome c oxidase. They block the binding site for oxygen in the active center.
proof
For the detection of the enzyme cytochrome c oxidase in cells is oxidase test used.
Alternative names
- Cytochrome c : O 2 oxidoreductase
- Cytochrome oxidase (cytochrome oxidase)
- Complex IV of the respiratory chain
- Cytochrome - aa 3 complex by David Keilin
- Respiratory ferment according to Otto Warburg (Nobel Prize for the discovery)
- Indophenol oxidase according to Paul Ehrlich
Other enzyme complexes in the respiratory chain:
- Complex I also NADH dehydrogenase
- Complex II also includes succinate dehydrogenase
- Complex III also cytochrome c reductase
- Complex V also ATP synthase
literature
- O. Warburg: About iron, the oxygen-transmitting component of the respiratory ferment. In: Biochemical Journal . Volume 152, 1924, pp. 479-494, doi: 10.1002 / cber.19250580603 .
- David Keilin: On cytochrome, a respiratory pigment, common to animals, yeast, and higher plants. In: Proc. Royal Soc. London. Series B. Volume 98, 1925, pp. 312-339, JSTOR 81121 .
- T. Tsukihara, H. Aoyama, E. Yamashita et al .: The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. In: Science . tape 272 , no. 5265 , May 1996, pp. 1136–1144 , doi : 10.1126 / science.272.5265.1136 , PMID 8638158 .
- S. Iwata, C. Ostermeier, B. Ludwig et al .: Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. In: Nature . tape 376 , no. 6542 , August 1995, p. 660-669 , doi : 10.1038 / 376660a0 , PMID 7651515 .
- F. Diaz: Cytochrome c oxidase deficiency: patients and animal models . In: Biochim Biophys Acta . tape 1802 , no. 1 , January 2010, p. 100-110 , doi : 10.1016 / j.bbadis.2009.07.013 , PMID 19682572 .
Individual evidence
- ↑ C. Orssaud: Optic neuropathy liver type. In: orpha.net. Orphanet, November 2003, accessed October 4, 2010 .
- ↑ Genetic recurrent myoglobinuria. In: orpha.net. Orphanet, March 2010, accessed October 4, 2010 .
- ↑ Mitochondrially inherited nonsyndromic sensorineural deafness. In: Online Mendelian Inheritance in Man . (English)
- ↑ 3.D.4 The Proton-translocating Cytochrome Oxidase (COX) Superfamily. In: TCDB. Saier Lab Bioinformatics, accessed October 4, 2010 .
- ^ YC Kim, M. Wikström, G. Hummer: Kinetic gating of the proton pump in cytochrome c oxidase. In: PNAS. 106/33, 2009, pp. 13707-13712, doi: 10.1073 / pnas.0903938106 .
- ↑ V. Sharma, G. Enkavi, I. Vattulainen, T. Róg, M. Wikström: Proton-coupled electron transfer and the role of water molecules in proton pumping by cytochrome c oxidase. In: PNAS. 112/07, 2015, pp. 2040–2045, doi: 10.1073 / pnas.1409543112 .
- ↑ T. Tsukihara, H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Nakashima, R. Yaono, S. Yoshikawa: Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. In: Science. 269, 5227, 1995, pp. 1069-1074, PMID 7652554 .
- ↑ B. Kadenbach , J. Jarausch, R. Hartmann, P. Merle: Separation of mammalian cytochrome c oxidase into 13 poly-peptides by a sodium dodecyl sulfate-gel electrophoretic procedure. In: Anal. Biochem. 129, 1983, pp. 517-521, PMID 6303162 .
Web links
- Jassal B: Electron transfer from reduced cytochrome c to molecular oxygen. In: reactome.org. EBI, June 28, 2005, accessed October 4, 2010 .