Orthomyxoviridae
| Orthomyxoviridae | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
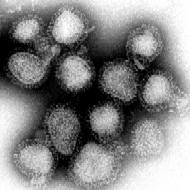
Influenza virus in the TEM |
||||||||||||||||
| Systematics | ||||||||||||||||
|
||||||||||||||||
| Taxonomic characteristics | ||||||||||||||||
|
||||||||||||||||
| Scientific name | ||||||||||||||||
| Orthomyxoviridae | ||||||||||||||||
| Left | ||||||||||||||||
|
The family Orthomyxoviridae ( Greek μύξα myxa , German 'slime' ) comprises enveloped viruses with single-stranded RNA with negative polarity as the genome . Their RNA genome is distributed over several segments, which is why, in contrast to the Paramyxoviridae, they are not assigned to the order Mononegavirales . The segmentation of their genome allows the Orthomyxoviridae a high genetic flexibility and adaptability to new host species due to the mixing of the different segments of different subtypes and mutants by the so-called reassortment .
The Orthomyxoviridae include virus genera that mainly infect the respiratory system of a host via droplet infection and multiply therein. This applies in particular to the genera of the influenza viruses , which can cause asymptomatic infections or serious illnesses in mammals and birds . Only the species of the genus Thogotovirus do not cause respiratory infections and are transmitted to vertebrates by ticks . Some aquatic hosts of the genera influenza virus A ( baleen whales ) and isavirus (salmon) are infected by contaminated water, direct contact or (in the case of the infectious salmon anemia virus ) by fish lice . The tilapia pond virus was also scientifically described as "orthomyxovirus-like" and placed by the ICTV in a new sister family Amnoonviridae , genus Tilapinevirus .
morphology
The virus particles (virions) of the Orthomyxoviridae are spherical to irregular and 80–120 nm in diameter. Thread-like (filamentous) shapes with lengths of up to a few µm are also observed. The lipid-containing virus envelope contains 1–3 glycoproteins and 1–2 non-glycosylated proteins. These form visible “spikes” on the surface that are 10–14 nm long and 4–6 nm in diameter. Anchored in the envelope, but with the greater part pointing inwards, are so-called matrix proteins that line the space between the envelope and the capsids (matrix space).
Depending on the segmentation of the genome, the virions also contain several helical capsids , at one end of which several subunits of the viral polymerase proteins (PA, PB1 and PB2) are associated. These viral enzymes show different activities depending on the virus genus, e.g. B. PB1 is an endonuclease in influenza viruses and also an RNA polymerase (transcriptase) in these and the genus Thogotovirus . Once released in the cytoplasm, the capsids are transported into the cell nucleus by specific nuclear import .
The (-) ssRNA genome is linear and segmented; the number of segments varies between the genera. Thus, the species of the genera Influenzavirus A , Influenzavirus B and Isavirus each have 8 segments, Influenzavirus C and from the genus Thogotovirus the species Dhori virus 7, the species Thogoto virus 6 segments. The size of the segments ranges from 874 to 2396 nt , the total size of the genome from 10.0 to 14.6 kb . Due to distribution and synthesis errors during virus replication, many virions have shorter, defective RNA pieces or do not have a complete set of segments. This is particularly observed in the case of mutations in the polymerase subunit PA, which apparently plays a decisive role in the correct packaging and distribution of the genome segments.
Systematics

As of March 2019, the systematics of the Orthomyxoviridae according to the International Committee on Taxonomy of Viruses (ICTV) is as follows:
-
- Family Orthomyxoviridae
-
- Genus alphainfluenza virus
- Species influenza virus A
- Genus betainfluenza virus
- Species influenza virus B
- Genus gamma fluenza virus
- Species influenza virus C
- Genus delta fluenza virus
- Species influenza virus D
- Genus Isavirus
- Species of Salmon Infectious Anemia Virus (officially Salmon isavirus )
- Genus Quaranja virus
- Species Quaranfil quaranjavirus ( Quaranfil virus , QRFV, type species)
- Species Johnston Atoll quaranjavirus ( Johnston Atoll Virus , JAV)
- Genus Thogotovirus
- Species Dhori virus
- Species Thogoto virus (officially Thogoto thogotovirus , type species including the so far unclassified ' Bourbon virus ')
The individual virus lines and other unclassified candidates can be found at the NCBI.
literature
- CM Fauquet, MA Mayo, et al. : Eighth Report of the International Committee on Taxonomy of Viruses. London, San Diego 2005.
- RA Lamb, CM Horvath: Orthomyxoviridae - The viruses and Their Replication. In: David M. Knipe, Peter M. Howley et al. (Ed.): Fields' Virology. 4th edition. Philadelphia 2001.
Web links
- Orthomyxoviridae: Genera and Species (NCBI)
- Image gallery of the Orthomyxoviridae family (Big Picture Book of Viruses)
swell
- ↑ a b ICTV: Master Species List 2018b.v2 , MSL # 34v, March 2019
- ↑ a b ICTV: ICTV Taxonomy history: Akabane orthobunyavirus , EC 51, Berlin, Germany, July 2019; Email ratification March 2020 (MSL # 35)
- ^ WR Kaufman, PA Nuttall: Rhipicephalus appendiculatus (Acari: Ixodidae): dynamics of Thogoto virus infection in female ticks during feeding on guinea pigs. Experimental Parasitology. 104 (1-2), May-June 2003, pp. 20-25.
- ↑ J. Mandler, OT Gorman, S. Ludwig, E. Schroeder, WM Fitch, RG Webster , C. Scholtissek: Derivation of the nucleoproteins (NP) of influenza A viruses isolated from marine mammals. Virology. 176 (1), May 1990, pp. 255-261.
- ↑ Eran Bacharach et al. : Characterization of a Novel Orthomyxo-like Virus Causing Mass Die-Offs of Tilapia. In: mBio. Volume 7, No. 2, e00431-16, 2016, doi: 0.1128 / mBio.00431-16
- ↑ G. Kochs, O. Haller: Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. PNAS 96 (5), March 2, 1999, pp. 2082-2086.
- ^ JF Regan, Y. Liang, TG Parslow: Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. Journal of Virology. 80 (1), Jan 2006, pp. 252-261. PMID 16352550 .
- ↑ NCBI: Orthomyxoviridae (family)