Paramyxoviridae
| Paramyxoviridae | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|

Measles virus in the thin-layer TEM |
||||||||||||||||
| Systematics | ||||||||||||||||
|
||||||||||||||||
| Taxonomic characteristics | ||||||||||||||||
|
||||||||||||||||
| Scientific name | ||||||||||||||||
| Paramyxoviridae | ||||||||||||||||
| Left | ||||||||||||||||
|
The Paramyxoviridae family includes enveloped viruses with a single-stranded, linear RNA with negative polarity as a genome . The Paramyxoviridae family originated from the outdated taxonomic group of myxoviruses (Greek myxa : slime) by differentiation (Greek para : next to) from the "real myxoviruses" ( Orthomyxoviridae ). The Paramyxoviridae include virus species that affect the respiratory system itself(e.g. the respiratory syncytial virus of the subfamily Pneumovirinae )or take their exit from it (species of the former subfamily Paramyxovirinae ). Therefore, Paramyxoviridae are not transmitted by vectors and almost exclusively by droplet infection . These are common and important diseases in mammals and birds, such as B. distemper and Newcastle disease , in humans it is infections such as measles , mumps and parainfluenza .
morphology
Virions
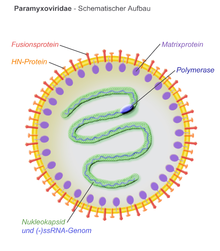
The virions of the Paramyxoviridae are usually round in shape and about 150 nm in diameter. Irregularly shaped or thread-like (filamentous) shapes are also described.
Virus envelope
The virus envelope comes directly from the outer cell membrane (similar to retroviruses ) and surrounds a helical capsid . 2 to 3 transmembrane glycoproteins are stored in the virus envelope . Several identical membrane proteins assemble into oligomers and form so-called spikes 8-12 nm in length and 7-10 nm apart (depending on the species). Outwardly, these form a fusion protein for penetrating the cell membrane and a protein for making contact ( attachment ) to the cell. A non-glycosylated membrane protein is always found with the Paramyxoviridae with its main part pointing inwards and thus lines the shell from the inside (so-called matrix proteins ).
The capsid (a ribonucleocapsid) is 13 to 18 nm thick and, depending on the length of the packed RNA strand, can be up to 1000 nm long in some species. Although multiple capsids can be found in one shell due to packaging errors during virus replication, there is mostly only one capsid and one RNA strand per virion . The virion also always contains at least one molecule of the viral RNA polymerase and sometimes parts of a complementary (+) ssRNA.
Genome
The viral genome consists of a single-stranded RNA with negative polarity . The length of the RNA is unusually constant and very similar within the genera. It is about 13 kB (genus metapneumovirus ) to 18 kB (genus Henipavirus ), with most paramyxoviruses mostly around 15 kB. If one takes a closer look at the length of individual members of the Paramyxovirinae subfamily , this follows a regularity that is unusual in viruses, which is divisible by the number 6: z. B. mumps virus 15,384 nt , Newcastle Disease Virus 15,156 nt. These multiples of the number 6 are due to a special mechanism of RNA replication in these viruses.
Biological properties
The Paramyxoviridae can infect mammals and birds . There are not yet classified viruses in fish (salmon of the genus Oncorhynchus ) and reptiles , but they are closely related to the paramyxoviruses . When they multiply, Paramyxoviridae dissolve the cell and are therefore cytolytic . The individual virus species are very closely adapted to their respective host and can therefore hardly be transferred from one host species to another. Paramyxoviridae usually cause an acute and self-limiting infection , which is eliminated again by an unaffected immune system of the host. A chronic or persistent infection is described as a special case only with measles virus (as subacute sclerosing panencephalitis ) and with canine distemper virus ( pathogen persistence in dogs and excretion over months).
Systematics

Internal system according to ICTV , as of November 2018 (excerpts):
- Family Paramyxoviridae
-
- (former) subfamily 'Paramyxovirinae'
-
- Genus Aquaparamyxovirus
- Salmon aquaparamyxovirus (aka Atlantic salmon paramyxovirus , AsaPV)
- Genus Avulavirus
- Avian avulavirus 1 - 19 (aka Avian paramyxovirus , APMV)
- Genus ferlavirus
- Reptilian ferlavirus (reptilian ferlavirus)
- Genus Henipavirus
- Cedar henipavirus (Cedar Virus, CedV)
- Hendra henipavirus ( Hendra virus , HeV, formerly equines morbillivirus ) - pneumonia ; Encephalitis
- Mojiang henipavirus (MojV)
- Nipah henipavirus ( Nipah virus , NiV) - pneumonia ; Encephalitis
- Genus Morbillivirus
- Canine morbillivirus (aka Canine distemper virus , CDV)
- Measles morbillivirus ( measles virus , MeV) - measles
- Rinderpest morbillivirus ( Rinderpestvirus , RPV)
- Small ruminant morbillivirus (aka Tupaia Paramyxovirus , TupPV)
- Genus Respirovirus
- Bovine respirovirus 3 (aka Bovine parainfluenza virus 3 , bPIV3)
- Human respirovirus 1 and 3 ( Human respirovirus 1 and 3, HPIV1 and… 3) - common cold , parainfluenza
- Murine respirovirus (aka Murine Parainfluenzavirus 1 , with Sendai virus )
- Genus Rubulavirus
- Human respirovirus 2 and 4 ( human parainfluenza virus 2 and 4, HPIV2 and… 4) - common cold , parainfluenza
- Mapuera rubulavirus ( Mapuera virus , MprPV)
- Menangle rubulavirus ( Menangle Virus , MenPV)
- Mumps rubulavirus ( mumps virus , MuV) - mumps
- Porcine respirovirus 1 (PorPV, swine rubulavirus)
- Tioman rubulavirus (alias Tioman paramyxovirus , Tioman virus , TioPV);
- From the second original subfamily Pneumovirinae within the family, the independent virus family Pneumoviridae emerged .
literature
- CM Fauquet, MA Mayo et al .: Eighth Report of the International Committee on Taxonomy of Viruses , London, San Diego 2005
- David M. Knipe, Peter M. Howley, et al. (eds.): Fields' Virology, 4th Edition, Philadelphia 2001
Web links
- Paramyxoviridae: Genera and Species (NCBI)
- Picture gallery of the Paramyxoviridae family (Big Picture Book of Viruses, via WebArchive from January 23, 2008)
Individual evidence
- ↑ ICTV Master Species List 2018b v1 MSL # 34, Feb. 2019
- ↑ a b ICTV: ICTV Taxonomy history: Akabane orthobunyavirus , EC 51, Berlin, Germany, July 2019; Email ratification March 2020 (MSL # 35)
- ↑ D. Kolakofsky, T. Pelet et al .: Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisted . Journal of Virology (1998) 197, 1-11
- ↑ J. Franke, S. Essbauer, W. Ahne, S. Blahak: Identification and molecular characterization of 18 paramyxoviruses isolated from snakes . Virus Research (2001) 28; 80: 67-74
- ↑ GA Marsh, C. de Jong, JA Barr, M. Tachedjian, C. Smith, D. Middleton, M. Yu, S. Todd, AJ Foord, V. Haring, J. Payne, R. Robinson, I. Broz , G. Crameri, HE Field, LF Wang: Cedar virus: a novel Henipavirus isolated from Australian bats . In: PLoS Pathogens . 8, No. 8, 2012, p. E1002836. doi : 10.1371 / journal.ppat.1002836 . PMID 22879820 . PMC 3410871 (free full text).
- ↑ ICTV : Master Species List 2018a v1 , MSL # 33 from autumn 2018.
- ↑ SIB: Avulavirus , on: ViralZone
- ↑ SIB: Respirovirus , on: ViralZone