Genome size
The genome size is the total amount of DNA in a copy of the genome .
properties
The genome size is given either in picograms , daltons or in mega base pairs . One picogram corresponds to 978 megabase pairs. In the case of diploid genomes, the size of the genome corresponds by convention to the C value , i.e. H. it is considered haploid .
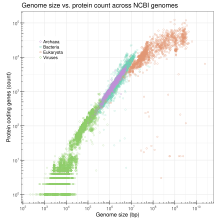
The C-value paradox describes the lack of a linear relationship between the amount of DNA and the complexity of a living being in eukaryotes , which results from a varying amount of non-coding DNA . There is some linear correlation in bacteria , archaea , viruses, and organelles , but not in eukaryotes. Furthermore, there is a certain correlation with the cell size, the rate of cell division and, within individual taxa , other parameters. There is also a connection between genome size and chromatin condensation . Further parameters for describing a genome are the gene density and the GC proportion . While there is a general tendency in the development of life to increase the genome size over time, the realms among the animals with the largest genomes are the fish with the lungfish (142 pg, corresponding to 139,000 MB) and the amphibians with the salamanders (83 pg , corresponding to 81,000 MB),
The genome size in animals varies by a factor of 3,300 and in plants by a factor of 2,300.
| Art | approximate genome size (in Mbp) | approximate number of genes | Gene density (in genes / Mbp) | Remarks |
|---|---|---|---|---|
| Porcine Circovirus-1 | 0.001759 | 2 | 1,137 | smallest genome of an autonomously replicating virus . |
| Carsonella Ruddii | 0.16 | 182 | 1,138 | smallest genome of a living being, endosymbiotic bacterium |
| Nanoarchaeum equitans | 0.49 | 536 | 914 | Archaea , parasitic |
| Mycoplasma genitalium | 0.58 | 800 | 860 | bacterium |
| Streptococcus pneumoniae | 2.2 | 2,300 | 1,060 | |
| Escherichia coli K12 | 4.6 | 4,400 | 950 | |
| Saccharomyces cerevisiae | 12.05 | 6.213 | 516 | yeast |
| Schizosaccharomyces pombe | 12 | 4,900 | 410 | yeast |
| Plasmodium falciparum | 22.85 | 5,268 | 231 | Malaria causative agent |
| Entamoeba histolytica | 23.75 | 9,938 | 418 | amoeba |
| Trypanosoma spp. | 39.2 | 10,000 | 255 | Flagellate |
| Tetrahymena thermophilus | 125 | 27,000 | 220 | Protist |
| Aspergillus nidulans | 30.07 | 9,541 | 317 | Mold |
| Neurospora crassa | 38.64 | 10,082 | 261 | Mold |
| Caenorhabditis elegans | 103 | 20,000 | 190 | Roundworm |
| Drosophila melanogaster | 180 | 14,700 | 82 | Fruit fly |
| Locusta migratoria | 5,000 | grasshopper | ||
| Takifugu rubripes | 393 | 22,000 | 56 | Puffer fish |
| Gallus gallus | 1,050 | 21,500 | 20.5 | chicken |
| homo sapiens | 3,200 | 20,000 | 6.25 | human |
| Mus musculus | 2,600 | 22,000 | 8.5 | House mouse |
| Arabidopsis thaliana | 120 | 26,500 | 220 | plant |
| Oryza sativa | 466 | 60,256 | 129 | rice |
| Zea mays | 2,200 | 45,000 | 20th | Corn |
| Picea glauca | 20,800 | 56.064 | 2.7 | Spruce |
| Lepidosis paradoxa | 78,400 | Lungfish | ||
| Protopterus aethiopicus | 139,000 | largest animal genome, lungfish |
The genome size is observed in a cell culture with a subsequent Feulgen reaction under a light microscope and counted using software or determined using flow cytometry .
For small genomes ( DNA viruses , unicellular cells ), John W. Drake postulated a reciprocal relationship between the mutation rate and the genome size in 1991 . In the case of RNA viruses , the even smaller genome size is a compromise between the mutation rate and the number of genes ( self-paradox ). The RNA polymerases of RNA viruses do not have an error correction function (no proof-reading ), which limits the size of the genome. The exception is the nidovirales , which have a proof-reading function with the exoribonuclease ExoN , which means that the genome size is less limited.
conversion
or changed:
Web links
Individual evidence
- ^ Eugene V. Koonin: The Logic of Chance: The Nature and Origin of Biological Evolution. FT Press, 2011. ISBN 9780132542494 .
- ↑ a b Dolezel J, Bartoš J, Voglmayr H, Greilhuber J, Bartos, Voglmayr, Greilhuber: Nuclear DNA content and genome size of trout and human . In: Cytometry Part A . 51, No. 2, 2003, pp. 127-128. doi : 10.1002 / cyto.a.10013 . PMID 12541287 .
- ^ S. Ohno: The Number of Genes in the Mammalian Genome and the Need for Master Regulatory Genes. In: Major Sex-Determining Genes. Monographs on Endocrinology. Volume 11. Springer, Berlin, 1979. ISBN 978-3-642-81261-3 . P. 17.
- ^ Y. Hou, S. Lin: Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: gene content estimation for dinoflagellate genomes. In: PloS one. Volume 4, number 9, September 2009, p. E6978, doi : 10.1371 / journal.pone.0006978 , PMID 19750009 , PMC 2737104 (free full text).
- ↑ Bennett MD, Leitch IJ: Genome size evolution in plants . In: TR Gregory (Ed.): The Evolution of the Genome . Elsevier, San Diego 2005, pp. 89-162.
- ↑ a b Gregory TR: Genome size evolution in animals . In: TR Gregory (Ed.): The Evolution of the Genome . Elsevier, San Diego 2005, pp. 3-87.
- ↑ a b Alexander E. Vinogradov: Genome size and chromatin condensation in vertebrates. In: Chromosoma. 113, 2005, p. 362, doi : 10.1007 / s00412-004-0323-3 .
- ↑ John Bernard: The Eukaryote Genome in Development and Evolution. Springer Science & Business Media, 2012, ISBN 978-9-401-15991-3 , p. 281.
- ↑ Johann Greilhuber: Plant Genome Diversity Volume 2. Springer Science & Business Media, 2012, ISBN 978-3-709-11160-4 , page 323rd
- ↑ National Center for Biotechnology Information : Porcine circovirus 1 isolate PCV1-Eng-1970, complete genome GenBank: KJ408798.1. In: Genbank . Retrieved January 24, 2018 .
- ↑ A. Nakabachi, A. Yamashita, H. Toh, H. Ishikawa, HE Dunbar, NA Moran, M. Hattori: The 160-kilobase genome of the bacterial endosymbiont Carsonella. In: Science. Volume 314, number 5797, October 2006, p. 267, doi : 10.1126 / science.1134196 , PMID 17038615 .
- ^ S. Das, S. Paul, SK Bag, C. Dutta: Analysis of Nanoarchaeum equitans genome and proteome composition: indications for hyperthermophilic and parasitic adaptation. In: BMC genomics. Volume 7, July 2006, p. 186, doi : 10.1186 / 1471-2164-7-186 , PMID 16869956 , PMC 1574309 (free full text).
- ↑ a b c d e f g h i j k l m James D. Watson: Molecular Biology. Pearson Deutschland GmbH, 2011, ISBN 978-3-868-94029-9 , pp. 172-175.
- ↑ a b c d e f g h Antonio Fontdevila: The Dynamic Genome. OUP Oxford, 2011, ISBN 978-0-199-54137-9 , p. 7.
- ^ I. Birol, A. Raymond, SD Jackman, S. Pleasance, R. Coope, GA Taylor, MM Yuen, CI Keeling, D. Brand, BP Vandervalk, H. Kirk, P. Pandoh, RA Moore, Y. Zhao , AJ Mungall, B. Jaquish, A. Yanchuk, C. Ritland, B. Boyle, J. Bousquet, K. Ritland, J. Mackay, J. Bohlmann, SJ Jones: Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. In: Bioinformatics. Volume 29, number 12, June 2013, pp. 1492-1497, doi : 10.1093 / bioinformatics / btt178 , PMID 23698863 , PMC 3673215 (free full text).
- ^ Northcutt RG. 1987. Lungfish neural characters and their bearing on sarcopterygian phylogeny. In The biology and evolution of lungfishes (ed. Bemis WE, et al.), Pp. 277-297. Alan R. Liss, New York.
- ↑ DC Hardie, TR Gregory, PD Hebert: From pixels to picograms: a beginners' guide to genome quantification by Feulgen image analysis densitometry. In: The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. Volume 50, Number 6, June 2002, pp. 735-749, doi : 10.1177 / 002215540205000601 , PMID 12019291 .
- ↑ JW Drake: A constant rate of spontaneous mutation in DNA-based microbes . In: Proc Natl Acad Sci USA . 88, 1991, pp. 7160-7164. doi : 10.1073 / pnas.88.16.7160 . PMID 1831267 . PMC 52253 (free full text).
- ^ A Kun, M Santos, E Szathmary: Real ribozymes suggest a relaxed error threshold . In: Nat Genet . 37, 2005, pp. 1008-1011. doi : 10.1038 / ng1621 . PMID 16127452 .
- ↑ C Lauber, JJ Goeman, C Parquet Mdel, P Thi Nga, EJ Snijder, K Morita, AE Gorbalenya: The footprint of genome architecture in the largest genome expansion in RNA viruses . In: PLoS Pathog . 9, No. 7, Jul 2013, p. E1003500. doi : 10.1371 / journal.ppat.1003500 .