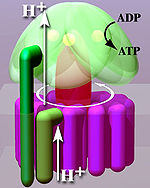
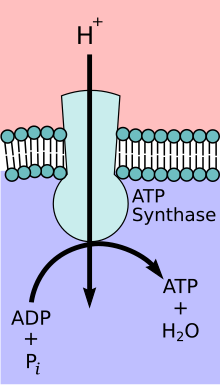
ATP synthase
| ATP synthase | ||
|---|---|---|
| Identifier | ||
| Gene name (s) | F o , F 1 | |
| Transporter classification | ||
| TCDB | 3.A.2 | |
| designation | F-ATPase superfamily | |
| Enzyme classification | ||
| EC, category | 3.6.3.14 , hydrolase | |
| Substrate | ADP + phosphate + H + outside | |
| Products | ATP + H 2 O + H + inside | |
The enzyme ATP synthase or F o F 1 -ATPase is a transmembrane protein . Depending on the ratio of substrates and products, the ATP synthase occurs either as an ATP-consuming proton pump or as a proton-driven ATP synthase. Under physiological conditions, however, the main task of the enzyme is to catalyze the synthesis of ATP . ATP is an energy-rich compound, the formation of which requires the supply of energy:
- ADP + phosphate → ATP + H 2 O with ΔH ≈ 30.5 kJ / mol under standard conditions and approx. 50 kJ / mol under physiological conditions.
In order to generate this energy, the ATP synthase couples the ATP formation with the energetically favored transport of protons (or other ions) along a proton gradient across a membrane. The ATP synthase is an energy converter that transforms one form of energy into another. The enzyme plays a central role in the metabolism of almost all known organisms, since ATP is continuously required as an energy carrier.
The ATP synthase is composed of 8 to 20 different subunits. They are grouped into two complexes:
- The water-soluble complex F 1 catalyzes the formation of ATP.
- The water-insoluble complex F o built into a membrane transports protons.
The enzyme is therefore also called F o F 1 -ATPase after its two subunits .
Meaning and occurrence
Practically all processes in organisms require adenosine triphosphate (ATP). It provides energy for all possible metabolic processes. Most of the ATP consumed is regenerated in animals, plants and most bacteria by the ATP synthase. The daily turnover of ATP in humans is sometimes well over 80 kilograms.
The ATP synthase alias F-type ATPase occurs
- in the plasma membrane of prokaryotes ("bacteria").
- in the inner mitochondrial membrane of eukaryotes (cells of plants and animals).
- in the thylakoid membrane of the chloroplasts of plant cells.
Primeval organisms from the archaea have an A-type ATPase, which differs somewhat from the "normal" ATP synthase in its structure. The reason could be related to the different structure of the cell membrane and cell wall of these organisms.
ATP synthases use the energy of an ion gradient that exists between the two sides of the membrane . As a rule, this is a proton gradient . In alkaliphilic bacteria there is also an ATP synthase that uses a sodium gradient for ATP synthesis instead of a proton. ( EC 3.6.3.15 )
history
The elucidation of the function and mechanism of ATP synthase was essentially done by researchers who dealt with mitochondria. Although this enzyme also plays an important role in photosynthesis in plants and in aerobic bacteria, the ATP synthase, as a component of cellular respiration and the human respiratory chain, became the focus of biochemistry .
The complex V
At the beginning of the 1960s, biochemistry could look back on enormous advances. The energy metabolism had almost been cleared up in a few decades. The citric acid cycle was known, as was the physiological role of NADH as a hydrogen carrier and the role of ATP as an energy supplier.
It was known that the NADH hydrogen is oxidized to water with oxygen. And it was clear that this process produced the bulk of the ATP required in the cell. In addition, it was known that the oxidation of NADH takes place in steps. Enzymes and coenzymes were found in the mitochondrial membranes, which form an electron transport chain from NADH to oxygen.
The elucidation of this so-called respiratory chain turned out to be increasingly difficult. The enzymes in the respiratory chain are difficult to isolate and investigate because they are membrane proteins . In addition, they form large enzyme complexes. Four of these complexes have been (and are) further elucidated. Complex V, which forms ATP, remained in the dark even after its water-soluble component was isolated in 1961. The elucidation of the respiratory chain, and with it the biochemistry in general, had one more "flaw".
From research on sugar breakdown ( glycolysis ), one already knew a mechanism ( substrate chain phosphorylation ) in which ADP and phosphate are converted into ATP. It was concluded that the respiratory chain must somehow behave similarly and it was believed to be close to the final clearing up. It was "only" missing
- the exact connection of the complexes I – IV to the synthesis of ATP, i.e. the coupling of O 2 consumption and ATP formation.
- the explanation of why the complex V found cleaved ATP but did not produce any once it had been isolated.
- an "energy-rich intermediate product" of the respiratory chain as a substrate for ATP synthase.
The only thing missing was the entire connection between NADH oxidation and ATP production. But one already had a name that is still valid today for the metabolic pathway: oxidative phosphorylation .
In this situation, Peter D. Mitchell presented a hypothesis in 1960 that was long and heavily attacked. Because Mitchell postulated a mechanism that was “unimaginable” for biochemistry at the time.
The Mitchell Hypothesis

Mitchell himself had focused on the respiratory chain since the early 1940s, but was also familiar with the research results of the transport processes on membranes. His research group concentrated on the electron carrier ubiquinone ( Q cycle ) when investigating the respiratory chain . The results were not compatible with the idea of an “energy-rich intermediate” in the respiratory chain as the motor of ATP synthase. Instead, Mitchell postulated that the enzyme gets its energy from a pH gradient .
The rejection of the professional world was overwhelming. Even when Mitchell received the Nobel Prize (chemistry) for his chemiosmotic theory in 1978 , well-known biochemists still spoke disparagingly of the "Mitchell hypothesis".
The elucidation of the mechanism
Even scientists who were officially very skeptical of the "Mitchell hypothesis" seriously considered the chemiosmotic theory in their research in the laboratory. While the evidence for Mitchell's theory increased, progress was made in the experimental use of ATP synthase. They could be studied "in action" in membrane vesicles .
Paul D. Boyer , originally a "Mitchell" skeptic, clarified the molecular mechanism of ATP synthase. John E. Walker and co-workers succeeded in crystallizing the ATP synthase and clarifying its spatial structure. They both received the 1997 Nobel Prize in chemistry. They shared the prize with Jens C. Skou , who discovered the first proton pump back in 1957 and thus laid the basis for the “Mitchell Hypothesis”.
Position on other enzymes
A large number of enzymes use the ATP supplied by the ATP synthase as a co- substrate . ATP synthase therefore plays a key role among enzymes.
The ATP synthase differs from the other ATPases in several ways:
- It is the main source of ATP. With its function as an ATP producer, the ATP synthase differs from the other (consistently) ATP-consuming ATPases, for example like an electricity generator from an electric motor.
- It consists of the two subunits F o and F 1 . Their function requires both units in a specific arrangement. The ATP synthase is therefore often referred to as F-type ATPase . In contrast, the ATPases that cleave ATP are V-type ATPases.
- Like all ATPases, the ATP synthase can in principle also function as a proton pump and thereby consume ATP. It is questionable, however, that this “reverse” reaction plays a major role in ATP synthase in vivo in mitochondria. The ATP synthase has a different “translation ratio” than the proton pump ATPases. The latter pump approx. Two protons outwards for every ATP consumed. In the case of ATP synthase, on the other hand, the energy of an ATP molecule would be distributed over three to four protons. As a proton pump, the ATP synthase could not build up such a large pH gradient .
In accordance with the IUBMB enzyme nomenclature, the ATP synthase has the EC number EC 3.6.3.14 and belongs to the hydrolase category .
ATP synthase blueprint
A distinction is made between a membrane-bound F o and a water-soluble F 1 subunit, depending on their position in relation to the membrane (Fig. Below, light area) .
From a mechanical point of view, the enzyme can be divided into a rotating rotor (Fig. “Simplified model”, reddish to purple) and a stator (Fig., Green). Due to the liquid nature of the membrane , the stator rotates in the opposite direction to the rotor.
Because of this structure, the enzyme, like the other membrane-bound ATPases, is a molecular machine .
The blueprint for the ATP synthase of the E. coli bacterium is described below because it has been studied intensively and is relatively simple.
Structure of F o
F o consists of hydrophobic peptides (water-insoluble protein chains) and is located in the membrane. This part of the enzyme is made up of three different subunits:
- F o a serves to transmit force to F o b and is at the same time part of the mechanism that converts the proton movement into a rotary movement.
- F o b connects the membrane and the F 1 component. F o b is used for power transmission and, like F o a, consists of two peptide chains .
- In E. coli, F o c consists of twelve spirals that are arranged in a ring. Inside the ring are probably the phospholipids from which the cell membrane is composed. They form an insulating layer so that no proton flow is possible there.
Each F o c peptide chain has an active center. If H + is split off from this center , the structure of the peptide chain changes into a mechanically tense state. If the active center then takes up an H + again , the peptide chain turns back. This rotation exerts a force on F o a.
Structure of F 1
F 1 is the water-soluble component of ATP synthase. It is located on the inside of the membrane. Five different peptides (α to ε) form the subunits of this component:
- Three F 1 α and F 1 β peptides each form the F 1 (αβ) 3 complex. Its designation as globular (αβ) 3 -ATPase , which is still used occasionally , indicates that the conversion of ADP to ATP takes place here. There are a total of three pores between the peptides through which the substrate and product can enter and exit. The ATP is produced in three catalytic centers inside the F 1 (αβ) 3 complex.
- F 1 γ is the rotatable axis of the system. It transmits the rotary motion of the ring placed in the membrane to the three catalytic (αβ) 3 centers.
- F 1 δ and F 1 ε are not shown in the figure on the right. The former peptide is the link between F o b and the (αβ) 3 complex. F 1 ε presumably connects the F o c ring with the F 1 γ axis.
Overview of human ATPase
Human ATPase consists of at least 16 subunits, two of which are encoded by mitochondrial genes. Several gene loci exist for both F 1 γ and F o c, which encode different precursors but identical subunits.
| Gene ( HGNC ) | Protein ( UniProt ) | Length ( AA ) | OMIM entry | Gene locus | comment |
|---|---|---|---|---|---|
| ATP5A1 | F 1 α1 | 553 | 164360 | 18q12-q21 | |
| ATP5B | F 1 β | 529 | 102910 | 12p13-qter | |
| ATP5C1 | F 1 γ1 | 298 | 108729 | 10q22-23 | |
| ATP5C2 | F 1 γ2 | ? | 14th | Isoform | |
| ATP5D | F 1 δ | 168 | 603150 | 19p13.3 | |
| ATP5E | F 1 ε | 51 | 606153 | 20q13.3 | |
| ATP5O | F 1 O | 213 | 600828 | 21q22.1-22.2 | Oligomycin sensitivity conferral protein |
| ATP5F1 | F o b | 256 | 603270 | 1p13.2 | |
| ATP5G1 | F o c isoform 1 | 136 | 603192 | 17q21.32 | Proteolipid P1; Subunit 9; Batten disease |
| ATP5G2 | F o c isoform 2 | 141 | 603193 | 12q13.3 | Proteolipid P2; Batten disease |
| ATP5G3 | F o c isoform 3 | 142 | 602736 | 2q31.1 | Proteolipid P3; Batten disease |
| ATP5H | F o d | 160 | 17q25 | stator | |
| ATP5I | F o e | 68 | 601519 | 4p16.3 | |
| ATP5J | F o F6 | 108 | 603152 | 21q21.1 | Coupling factor 6 |
| ATP5J2 | F o f | 93 | 7q22.1 | ||
| ATP5L | F o g | 103 | 11q23.3 | ||
| ATP5S | F o s | 215 | 14q21.3 | Coupling factor B |
Movement and reaction sequences
The function of the “molecular machine” is described here using the example of E. coli -ATP synthase.
Hypothetical mechanism of rotation
The location of the center responsible for the movement is shown in the figure " Motor ". The mechanism of the rotary movement is shown as an animation in the figure on the right.
|
Each F o c peptide from the rotor ring has an aspartic acid (ASP) residue at position 61 . When the engine is idle, the ASP carboxyl groups are protonated (peptide COOH) with one exception. In the rotor peptide 1, which is located next to the stator F o b, the ASP group is located next to an arginine group of the stator. Their positive charge can stabilize a negative charge through ionic interaction, so that the ASP residue is deprotonated (peptide-COO - ). The negatively charged rotor peptide 1 differs in its spatial structure from the other F o c peptides. In a hairpin loop , like in a spiral spring , it has built up a strong mechanical tension . |
|
|
Rotor peptide 1 picks up a proton from outside:
(Which path the proton takes has not yet been precisely clarified. The F o c peptides may create the space for it to penetrate to the decisive position 61. According to the "single channel theory", the stator peptide creates the access.) |
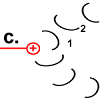
|
As soon as rotor peptide 1 is protonated, its tension-rich structure returns to the relaxed normal state. The "spiral spring" releases its energy again. A force is exerted on the stator. The peptide rotates.
Chemical energy is converted into kinetic energy here. The rotation of peptide 1 drives the motor. |

|
The rotary motion brings peptide 1 to the other side of the stator arginine and rotates the rotor ring. |
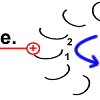
|
Rotor peptide 1 now has the same shape as the other rotor peptides that have no stator as a neighbor: protonated and with a relaxed spiral structure. |

|
The ring is now rotated by 30 °.
The movement of the rotor peptide, however, at the same time pulled the next F o c peptide chain 2 under the “spell” of the positively charged arginine group. |
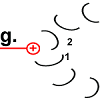
|
The structure of peptide 2 is changed to the energized state. The "spiral spring" is tensioned. |

|
Peptide 2 separates from the H + it had on the aspartate group at position 61. The proton leaves the location of the action towards the inside (in the picture: upwards).
|

|
The initial state is restored. However, it was
|
ATP synthesis in the F 1 (αβ) 3 complex
From a biochemical point of view, the actual conversion of ADP into ATP is nothing unusual compared to the proton-driven rotation of the F o c ring. There are structural changes in the peptide chains involved, which cause the substrates (here ADP and phosphate) to react.
There are three catalytic centers in the F 1 (αβ) 3 complex. They take three forms in sequence:
- High affinity for ADP and phosphate. The starting products ADP and phosphate are bound to the respective catalytic center.
- High affinity for ATP. The center takes on a hydrophobic character, which energetically favors the condensation reaction to form ATP.
- Open form in which ATP is expelled.
The energy-consuming step is to form the opened shape, i.e. to remove the reaction product ATP from the enzyme. This is exactly what the rotary movement enables.
The rotation of the rotor by 360 ° delivers three molecules of ATP in three steps. Since the “machine” rotates by 30 ° with each proton passage, four H + are consumed for each ATP according to this model .
ATP synthases in various organisms
The ATP synthase of the E. coli bacterium consists of eight different proteins. The ATP synthase in the mitochondria of the yeast Saccharomyces cerevisiae consists of 20 different proteins with basically the same structure.
There is also a wide range (9–14) in the number of F o c rotor peptides. They result in different proportions between H + consumed and ATP produced.
| Type / origin | Number of F o c peptides | H + per ATP |
|---|---|---|
| Bacteria ( E. coli ) | 10 | 4th |
| Mitochondria (yeast) | 10 | 3.3 |
| Chloroplasts (spinach) | 14th | 4th |
| Na + ATP synthase | 11 | 3.7 Na + / ATP |
Other names for the ATP synthase
- ATP synthetase
- Strictly speaking, this is an incorrect designation, as synthetases create a chemical bond while consuming ATP.
- ATPase
- The ATP synthase is the most common ATPase . Therefore, both names are often used synonymously.
- F-ATPase, F o F 1 -ATPase (or F 1 F o -ATPase), H + -ATP-synthase, H + -transporting two-sector ATPase
- ATP synthase in bacteria, mitochondria and plastids: Formally correct, but sometimes less common names
- A-ATPase, A o A 1 -ATPase
- ATPase in archaea is similar in structure to V-ATPase in eukaryotic vesicles. Occasional exceptions (A-type in certain bacteria, F-type in individual archaea species) are attributed to horizontal gene transfer .
- F zero F 1 - alias F 0 F 1 -ATPase, A 0 A 1 -ATPase, V 0 V 1 -ATPase, FoFI ATP-
- Incorrect. The name of the F o subunit is derived from oligomycin . This is a poison ( inhibitor ) that blocks this subunit.
- mtATP synthase
- ATP synthase in mitochondria
- CF o F 1 -ATP synthase
- ATP synthase in chloroplasts
The direction in which an ATP synthase acts can also be reversed according to the chemical-osmotic equilibrium - the internal motor activity of the enzyme does not change anything (see DC machine : depending on the direction of the power flow: dynamo versus electric motor). The ATPases then work as real membrane-bound synthetases. Examples are V-ATPase, N-ATPase, P-ATPase (the latter with a different structure, without rotating parts) and E-ATPase. More information about the different types can be found under membrane-bound ATPases, section: types .
literature
- PD Boyer, RL Cross, W. Momsen: A new concept for energy coupling in oxidative phosphorylation based on a molecular explanation of the oxygen exchange reactions. In: Proceedings of the National Academy of Sciences . Volume 70, Number 10, October 1973, pp. 2837-2839, PMID 4517936 , PMC 427120 (free full text).
- Armen Y Mulkidjanian, Kira S Makarova, Michael Y Galperin, Eugene V Koonin: Inventing the dynamo machine: the evolution of the F-type and V-type ATPases . In: Nature Reviews Microbiology . 5, No. 11, 2007, pp. 892-899. doi : 10.1038 / nrmicro1767 . This article (PDF) at Uni Osnabrück: Perspectives
- Armen Y Mulkidjanian, Michael Y Galperin, Eugene V Koonin: Co-evolution of primordial membranes and membrane proteins . In: Trends Biochem Sci . 4, No. 34, 2009, pp. 206-215. doi : 10.1016 / j.tibs.2009.01.005 . PMC 2752816 (free full text).
- Nick Lane: The Spark of Life - Energy and Evolution . Konrad Theiss Verlag, 2017 by WBG, ISBN 978-3-8062-3484-8 . Original English Title: Nick Lane: The Vital Question - Energy, Evolution, and the Origins of Complex Life . Ww Norton, 2015, ISBN 978-0-393-08881-6 , armscoop.com (PDF). Text passages near Figure 10 (structure of the ATP synthase).
- E. Hilario, JP Gogarten: Horizontal transfer of ATPase genes - the tree of life becomes a net of life . In: Biosystems , 1993, 31 (2-3), pp. 111-119. PMID 8155843
Web links
- Paul D. Boyer: ATP Synthase: A Magnificent Molecular Machine .
- BA Feniouk: ATP Synthase FAQ . (English)
- Olaf Fritsche: Biophysicists discovered the oldest wheel in the world .
- P. Gräber, A. Labahn: The H + -ATPsynthase . (German)
- Nobel Prize in Chemistry 1997 presentation. Nobel Media AB
- Kanehisa Laboratories (KEGG): Oxidative phosphorylation - Reference pathway . (with graphic representation in connection with the respiratory chain and links to the EC numbers)
- TCDB: View Proteins belonging to: The H + - or NaH + -translocating F-type, V-type and A-type ATPase (F-ATPase) Superfamily . (English)
- Jennifer McDowall / Interpro: Protein of the Month: ATP Synthase. (English)
- Jassal / Reactome: Formation of ATP by chemiosmotic coupling .
Individual evidence
- ↑ a b S. Steigmiller, P. Turina, P. Gräber: The thermodynamic H + / ATP ratios of the H + -ATPsynthases from chloroplasts and Escherichia coli. In: Proceedings of the National Academy of Sciences . Volume 105, number 10, March 2008, pp. 3745-3750, doi: 10.1073 / pnas.0708356105 , PMID 18316723 , PMC 2268821 (free full text).
- ↑ Elena Hilario, Johann Peter Gogarten: Horizontal transfer of ATPase genes - the tree of life becomes a net of life . In: BioSystems . 31, No. 2-3, 1993, pp. 111-119. doi : 10.1016 / 0303-2647 (93) 90038-E . PMID 8155843 ScienceDirect University of Connecticut (PDF)
- ^ G. Grüber et al .: ATP synthases from archaea: The beauty of a molecular motor . In: Biochimica et Biophysica Acta (BBA) - Bioenergetics . Volume 1837, Issue 6, 2014, pp. 940–952, doi: 10.1016 / j.bbabio.2014.03.004 (English).
- ↑ Jennifer McDowall: ATP Synthase: The ATPase-Family (English)
- ^ AK Srivastava, NK Ramaswamy, R. Mukopadhyaya, MG Jincy, SF D'Souza: Thiourea modulates the expression and activity profile of mtATPase under salinity stress in seeds of Brassica juncea. In: Annals of botany. Volume 103, number 3, February 2009, pp. 403-410, doi: 10.1093 / aob / mcn229 , PMID 19033283 , PMC 2707324 (free full text).