Tomato bushy stunt virus
| Tomato bushy stunt virus | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|

The capsid of Tomato bushy-stunt virus |
||||||||||||||||||||
| Systematics | ||||||||||||||||||||
|
||||||||||||||||||||
| Taxonomic characteristics | ||||||||||||||||||||
|
||||||||||||||||||||
| Scientific name | ||||||||||||||||||||
| Tomato bushy stunt virus | ||||||||||||||||||||
| Short name | ||||||||||||||||||||
| TBSV | ||||||||||||||||||||
| Left | ||||||||||||||||||||
|
The Tomato bushy stunt virus ( scientifically Tomato bushy stunt virus , TBSV ), rarely also referred to as tomato dwarf bush virus , is the type species of the virus genus Tombusvirus from the family Tombusviridae , subfamily Procedovirinae . TBSV is a single stranded RNA virus that was first found in tomatoes in 1935. It primarily affects vegetable crops but is not generally considered to be an economically significant plant pathogen . Depending on the host , TBSV causes growth disorders , leaf spots and deformed or missing fruits. The virus is likely to be grounded in nature, but it can also be transmitted mechanically, for example through contaminated cutting tools. TBSV has been used as a model system in virological research on the life cycle of plant viruses, especially in experimental infections of the model host plant Nicotiana benthamiana .
The stunted ( English to stunt , 'wither' ), "bushy" appearance of the tomato plants in which the virus was first discovered, gave the pathogen its name.
Host plants
TBSV is a broad host spectrum under experimental conditions and has been reported to infect over 120 different plant species from 20 families. However, in natural conditions, its range is much narrower and generally includes harvested vegetables and ornamentals . It was first detected in tomato plants, but has also been documented as a parasite of apples , artichokes , cherries , grapevines , hops , pepper and wild strawberries . Although TBSV leads to significant yield losses in tomato plants, it is not considered an economically important pathogen. However, it is a very well established model system for studying plant viruses, usually by experimental infection of Nicotiana benthamiana or N. clevelandii , relatives of the tobacco plant in which TBSV can cause systemic infection. However, the common model plant Arabidopsis thaliana is not a host. Interestingly, TBSV can also replicate in yeast (brewer's yeast Saccharomyces cerevisiae ) under laboratory conditions .
Infection and its signs
The signs of TBSV are host dependent. Local infections can cause necrotic or chlorotic lesions . Systemic infections can lead to stunted growth , deformed or missing fruits, and damaged leaves such as the stunted, "bushy" appearance of tomato plants. In agriculture, the yield can be significantly reduced. In some hosts, particularly N. benthamiana , TBSV can cause fatal systemic necrosis.
transmission
It is believed that TBSV is passively transmitted in the wild, primarily through the soil and water. No vector organisms are known, in particular transmission by aphids, mites or the droplet fungus Olpidium brassicae ( Olpidiaceae ) is expressly excluded. However, it has been observed that the closely related Cucumber necrosis virus (CNV), also virus genus Tombusvirus , is transmitted by zoospores of the closely related droplet species O. bornovanus , so that the transmission of TBSV by a previously unknown vector is still possible. TBSV can also be transmitted by seed or by mechanical inoculation (inoculation). In experimental tests, the virus can survive passage through the human digestive system if ingested in food and remains contagious. It is therefore believed that it could spread through the sewage.
Dissemination and countermeasures
TBSV is fairly widespread in Central and Western Europe, North Africa, and North and South America. There are no recommendations for specific control measures. However , the pest control guidelines issued by the University of California recommend avoiding fields with a history of TBSV or using long crop rotations .
Systematics
TBSV is the type species of the genus Tombusvirus in the family Tombusviridae , subfamily Procedovirinae . Both the genus and the family take their name from an abbreviation for English tomato bushy stunt virus derived.
- Family Tombusviridae
-
- Subfamily Procedovirinae
-
- Genus Tombus Virus
-
- Species Carnation Italian ringspot virus
- Species Cucumber Bulgarian latent virus
- Species Cucumber necrosis virus (en. Cucumber necrosis virus , CNV)
- Species Cymbidium ringspot virus
- Species Eggplant mottled crinkle virus
- Species Grapevine Algerian latent virus
- Species Havel River virus
- Species Lato River virus
- Species Limonium flower distortion virus
- Species Moroccan pepper virus
- Species Neckar River virus
- Species Pelargonium leaf curl virus
- Species Pelargonium necrotic spot virus
- Species Petunia asteroid mosaic virus
- Species Sitke waterborne virus
- Species Tomato bushy stunt virus ( en.Tomato bushy stunt virus , TBSV, type species), isolates according to National Center for Biotechnology Information (NCBI)
- TBSV strain BS-3
- TBSV strain CHERRY
- TBSV strain A23
- TBSV strain b8
- TBSV strain yes 6
- TBSV strain yes 9
- TBSV strain type
construction
The virus particles ( virions ) of TBSV are not enveloped and have an icosahedral geometry ( T = 3 symmetry ). The capsid is composed of 180 subunits of a single capsid protein. Its structure was extensively studied by X-ray crystallography from the late 1950s . The icosahedral symmetry was first identified by the structural biologist Donald Caspar, who also did pioneering work in researching the tobacco mosaic virus . In 1978 a research team led by Stephen C. Harrison achieved a representation with almost atomic resolution.
Genome and proteome
TBSV is a single-stranded RNA virus with positive polarity and a linear genome about 4800 nt in length . The RNA strand lacks the 3 'polyadenine tail and the 5' cap .
The genome contains five genes that encode a replicase , which is composed as follows:
- two proteins (p33 and p92),
- a capsid protein (called CP or p41),
- two other proteins: the RNA silencing suppressor p19 and the movement protein p22,
- maybe there is another gene of unknown function called pX.
The two proteins p19 and p22 are expressed from overlapping genes which are arranged so that the open reading frame ( English open reading frame , ORF) of p19 is completely in the ORF of p22.
p33 and p92
The two proteins p33 and p92 together form the viral replicase complex. P33 is smaller than p92. The latter is generated by ribosomal reading of the p33 stop codon, which leads to a common N-terminal amino acid sequence and a large excess of p33 compared to p92. The p33 proteins cooperatively bind single-stranded nucleic acids, while the p92 protein is an RNA-dependent RNA polymerase (RdRp). Both are essential for virus proliferation (the multiplication of virus particles). Both proteins are associated with cell membranes.
p41 (capsid protein)
The capsid protein p41 (alias CP) is a double jelly roll protein ( English jelly roll fold ). The icosahedral capsid is composed of 180 copies of the protein. The formation of virions is not absolutely necessary in order to locally spread the virus into neighboring plant cells, since ribonucleoprotein particles with the viral genetic material can spread to the immediate neighboring cells via plasmodesmata (plasmodesmata). For a systemic infection (of other plants) by fully functional virions, however, the capsid protein is required.
p19
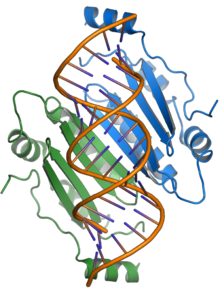
The p19 protein is a pathogenicity factor and suppresses the RNA silencing signaling pathway, a common form of antiviral defense on the part of the hosts. The p19 protein binds short sRNAs (“small RNAs”) and prevents their incorporation into the RNA-induced silencing complex (RISC), which enables the virus to multiply in the host plant. The presence of p19 is required for systemic or fatal infection in some hosts; in the experimental host N. benthamiana , lethal systemic necrosis as a result of TBSV infection is largely mediated by p19.
p22 (movement protein)
The p22 protein is a movement protein that is required for the virus to spread from cell to cell. It is an RNA-binding protein that is associated with the cell wall and facilitates the movement of viral genetic material from one cell to its neighbor through connective plasmodesmata .
Replication
Although the RNA strand of the genome of the 3 'poly A tail and the 5' cap is missing, the proteins p33 and p92 directly from genomic RNA translated . When the genome is replicated, two subgenomic RNA molecules are created that function as messenger RNA ( mRNA ): one from which the capsid protein p41 (CP) is expressed and one from which the proteins p19 and p22 are expressed. The overlapping genes p19 and p22 are both translated via the effects of leaky scanning . In addition, several interactions between linearly well separated and widely spaced regions of the genome were identified with functional importance for efficient replication.
DI particles
DI molecules ( English defective interfering RNA ) are prepared from the viral genome, but which cells are unable due to shortening and other defects, (such as a virus) infecting RNAs alone. Instead, like a satellite virus, they must be co-infected with an intact helper virus . TBSV infections often produce a significant number of DIs from complete and functional parts of the genome under experimental conditions; however, this behavior has not been observed in the wild. Your production is likely to be host specific. Infections that lead to DIs are usually milder.
siRNAs
A variant of the small sRNAs is the siRNA ( English small interfering RNA ). This occurs when the plant recognizes and fragmented virus genes. Together with certain proteins (signpost or helper proteins) of the plant, these can bind to the virus RNA and inactivate it to protect the plant; an effect that is only possible with a very small number of siRNAs. Attempts made in 2019 to inject suitable siRNAs into the N. benthamiana plants with the help of Agrobacterium tumefaciens , and thus to effect a kind of immunization of the plants, were promising. These bacteria are able to overcome the cell wall of the plant cells and penetrate inside.
Web links
- Tomato Bushy Stunt Virus , on ScienceDirect
Individual evidence
- ^ P. Hopper, SC Harrison, RT Sauer: Structure of tomato bushy stunt virus. V. Coat protein sequence determination and its structural implications. . In: Journal of Molecular Biology . 177, No. 4, August 25, 1984, pp. 701-713. doi : 10.1016 / 0022-2836 (84) 90045-7 . PMID 6481803 .
- ↑ a b c ICTV Master Species List 2018b v2 MSL # 34v, March 2019
- ↑ a b c d ICTV: ICTV Taxonomy history: Carrot mottle mimic virus , EC 51, Berlin, Germany, July 2019; Email ratification March 2020 (MSL # 35)
- ↑ a b Penka Kaitasowa, Violeta Kondakowa: Immunoelectron microscopic detection of the tomato bushy stunt virus in strawberries: (short communication) , in: Arch. Phytopathol. Plant protection, Berlin December 20, 1988, doi: 10.1080 / 03235408909438914 , online September 12, 2009
- ^ Brian WJ Mahy, Marc HV Van Regenmortel: Desk Encyclopedia of Plant and Fungal Virology . Academic Press, October 15, 2009, ISBN 978-0-12-375148-5 , pp. 445– (accessed August 26, 2019).
- ^ A b c Herman B. Scholthof: The Tombus virus-encoded P19: from irrelevance to elegance . In: Nature Reviews Microbiology . 4, No. 5, March 6, 2006, pp. 405-411. doi : 10.1038 / nrmicro1395 . PMID 16518419 .
- ↑ a b c d e f g h i j k l m n o Y. Yamamura, HB Scholthof: Tomato bushy stunt virus: a resilient model system to study virus-plant interactions . In: Molecular Plant Pathology . 6, No. 5, September 1, 2005, pp. 491-502. doi : 10.1111 / j.1364-3703.2005.00301.x . PMID 20565674 .
- ↑ a b c d e GP Martelli, M. Russo, L. Rubino: Tomato bushy stunt virus . Association of Applied Biologists. December 2001. Retrieved August 26, 2019.
- ↑ Fanny Balique, Hervé Lecoq, Didier Raoult, Philippe Colson: Can Plant Viruses Cross the Kingdom Border and Be Pathogenic to Humans? . In: Viruses . 7, No. 4, April 20, 2015, pp. 2074-2098. doi : 10.3390 / v7042074 . PMID 25903834 . PMC 4411691 (free full text).
- ^ Integrated Pest Management Program . University of California Division of Agriculture and Natural Resources. December 2013. Retrieved August 26, 2019.
- ^ ICTV Taxonomy History for Tomato bushy stunt virus . July 2015. Retrieved December 16, 2016.
- ^ BD Harrison, JT Finch, AJ Gibbs, M. Hollings, RJ Shepherd, V. Valenta, C. Wetter: Sixteen groups of plant viruses . In: Virology . 45, No. 2, August 1, 1971, pp. 356-363. doi : 10.1016 / 0042-6822 (71) 90336-9 .
- ↑ NCBI : Tomato bushy stunt virus (species)
- ^ Michael G. Rossmann: Structure of viruses: a short history . In: Quarterly Reviews of Biophysics . 46, No. 2, May 1, 2013, ISSN 0033-5835 , pp. 133-180. doi : 10.1017 / S0033583513000012 . PMID 23889891 .
- ^ SC Harrison, AJ Olson, CE Schutt, FK Winkler, G. Bricogne: Tomato bushy stunt virus at 2.9 Å resolution . In: Nature . 276, No. 5686, November 23, 1978, pp. 368-373. doi : 10.1038 / 276368a0 . PMID 19711552 .
- ↑ Alexander Hellemans, Bryan H. Bunch: The timetables of science: a chronology of the most important people and events in the history of science . Simon and Schuster, 1988, ISBN 978-0-671-62130-8 , p. 656 (accessed August 26, 2019).
- ↑ Patrick Q. Hearne, David A. Knorr, Bradley I. Hillman, Thomas J. Morris: The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus . In: Virology . 177, No. 1, July 1, 1990, pp. 141-151. doi : 10.1016 / 0042-6822 (90) 90468-7 .
- ^ Edward K. Wagner, Martinez J. Hewlett, David C. Bloom, David Camerini: Basic Virology . John Wiley & Sons, November 6, 2007, ISBN 9781405147156 , pp. 268– (Accessed August 26, 2019).
- ↑ a b Baodong Wu, Jörg Grigull, Moriam O. Ore, Sylvie Morin, K. Andrew White: Global Organization of a Positive-strand RNA Virus Genome . In: PLOS Pathogens . 9, No. 5, May 23, 2013, ISSN 1553-7374 , p. E1003363. doi : 10.1371 / journal.ppat.1003363 . PMID 23717202 . PMC 3662671 (free full text).
- ↑ a b Herman B. Scholthof, Karen-Beth G. Scholthof, Marjolein Kikkert, AO Jackson: Tomato Bushy Stunt Virus Spread Is Regulated by Two Nested Genes That Function in Cell-to-Cell Movement and Host-Dependent Systemic Invasion . In: Virology . 213, No. 2, 1995, pp. 425-38. doi : 10.1006 / viro.1995.0015 . PMID 7491767 .
- ↑ Richard W. Jones, AO Jackson, Thomas J. Morris: Defective-interfering RNAs and elevated temperatures inhibit replication of tomato bushy stunt virus in inoculated protoplasts . In: Virology . 176, No. 2, 1990, pp. 539-45. doi : 10.1016 / 0042-6822 (90) 90024-L . PMID 2345965 .
- ↑ Tomato bushy stunt virus (TBSV), p22, movement protein (IPR005332) - Short name: TBSV_p22 , to: InterPro - Protein sequence analysis & classification
- ↑ shorter than the (RNA) genome
- ↑ Karen-Beth G. Scholthof, Herman B. Scholthof, Andrew O. Jackson: The Effect of Defective Interfering RNAs on the Accumulation of Tomato Bushy Stunt Virus Proteins and Implications for Disease Attenuation . In: Virology . 211, No. 1, August 1, 1995, pp. 324-328. doi : 10.1006 / viro.1995.1410 . PMID 7645230 .
- ↑ NCBI: Defective interfering RNA-4 of tomato bushy stunt virus (TBSV-P DI-4) and Defective interfering RNA-5 of tomato bushy stunt virus (TBSV-P DI-5)
-
↑
Selma Gago-Zachert, Jana Schuck, Claus Weinholdt, Marie Knoblich, Vitantonio Pantaleo, Ivo Grosse, Torsten Gursinsky, Sven-Erik Behrens: Highly efficacious antiviral protection of plants by small interfering RNAs identified in vitro ;
Claus Weinholdt, Ivo Grosse, Jana Schuck, Sven-Erik Behrens, Vitantonio Pantaleo, Torsten Gursinsky: Highly efficacious antiviral protection of plants by small interfering RNAs identified in vitro ;
in: Nucleic Acids Research, August 21, 2019, doi: 10.1093 / nar / gkz678 - ↑ MLU: Plant protection: researchers develop new modular vaccination kit , press release number 128/2019 from Martin Luther University Halle-Wittenberg from August 21, 2019
- ↑ Annika Röcker: Selected vaccines against plant killers , on: Spektrum.de from August 22, 2019
