The Satanic Bible and User:Marshallsumter: Difference between pages
m Reverted edits by 71.17.86.207 to last version by 24.87.145.12 (HG) |
|||
| Line 1: | Line 1: | ||
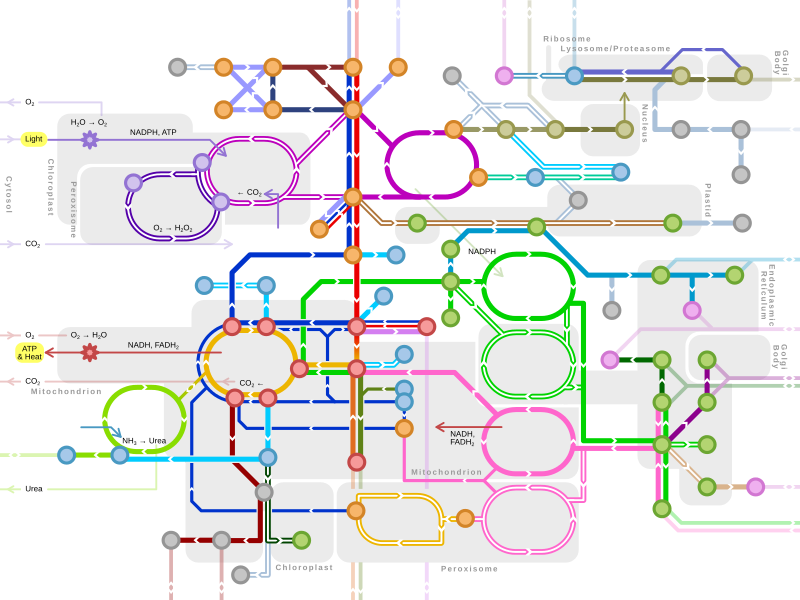
[[Biosynthesis]] of a [[Human]] [[Protein]] |
|||
{{Infobox Book |
|||
| name = The Satanic Bible |
|||
{{Taxobox |
|||
| title_orig = |
|||
| color |
|||
| translator = |
|||
| name = [[Human]] Lineage |
|||
| image = [[Image:satanicbible.gif|150px|The Satanic Bible]] |
|||
| domain = [[Eukaryote|Eukaryota]] |
|||
| image_caption = |
|||
| |
| regnum = [[Animal]]ia|Metazoa]] |
||
| phylum = [[Chordate|Chordata]] |
|||
| illustrator = |
|||
| subdivisio = [[Craniata]] |
|||
| cover_artist = |
|||
| |
| subphylum = [[Vertebrate|Vertebrata]] |
||
| infraphylum = [[Euteleostomi]] |
|||
| language = [[English language|English]] |
|||
| classis = [[Mammal|Mammalia]] |
|||
| series = |
|||
| infraclassis = [[Eutheria]] |
|||
| genre = [[Non-fiction]], [[Satanism]], [[Philosophy]] |
|||
| superordo = [[Euarchontoglires]] |
|||
| publisher = |
|||
| ordo = [[Primate|Primates]] |
|||
| release_date = 1969 |
|||
| subordo = [[Haplorrhini]] |
|||
| media_type = Print ([[Hardcover|Hardback]] & [[Paperback]]) |
|||
| infraordo = [[Simiiformes]] |
|||
| pages = |
|||
| parvordo = [[Catarrhini]] |
|||
| isbn = |
|||
| parvordo_authority = [[Étienne Geoffroy Saint-Hilaire|É. Geoffroy]], 1812 |
|||
| followed_by = [[The Satanic Rituals]] |
|||
| familia = [[Great ape|Hominidae]] |
|||
| subfamilia = [[Homininae]] |
|||
| tribus = [[Hominini]] |
|||
| subtribus = [[Hominina]] |
|||
| genus = [[Homo (genus)|Homo]] |
|||
| genus_authority = [[Carolus Linnaeus|Linnaeus]], 1758 |
|||
| species = sapiens |
|||
| subspecies = sapiens |
|||
''[[Human|Homo sapiens]] sapiens''<br> |
|||
}} |
}} |
||
{{Satanism}} |
|||
'''''The Satanic Bible ''''' was written by [[Anton LaVey]] in 1969. It is a collection of essays, observations and basic Satanic [[ritual]]s, and outlines LaVey's Satanic ideology. The author claims the influence of [[Ragnar Redbeard]] and [[Ayn Rand]] among others. |
|||
The book contains the core principles of [[Satanism]] and is considered the foundation of the philosophy and dogma that constitute Satanism. [[LaVeyan Satanism|LaVeyan Satanists]] maintain that ''The Satanic Bible'' may not be subject to interpretation or revision, and that no rule or principle contradictory to what is written in ''The Satanic Bible'' may be considered applicable to Satanism.{{Fact|date=February 2008}} |
|||
Consider starting with a specific gene. Assemble the transcription mechanism at the appropriate genome location. Transcribe the gene using nucleotide accountability. |
|||
==History== |
|||
''The Satanic Bible'' was written in 1969 by [[Anton Szandor LaVey]] and has been in continuous print ever since. The main text (original English) has never been altered in any edition; however, the full dedication was removed following LaVey's divorce from his wife Diane. Introductions to the work have been added and replaced. Burton H. Wolfe, a journalist who wrote ''The Devil's Avenger'', wrote the introduction to the original edition and later softcover printings, while [[Michael Aquino]] contributed the introduction to the hardcover edition published by [[University Press of America|University Press]].{{Fact|date=September 2007}} This introduction is reprinted in Aquino's [http://www.xeper.org/maquino/nm/COS.pdf ''Church of Satan''] memoir. The current Spokesman and Second High Priest of the Church of Satan, [[Peter H. Gilmore]], wrote a new introduction that replaced Wolfe's and was included with The Satanic Bible starting in 2005. University Press released a hardcover edition of The Satanic Bible and [[The Satanic Rituals]], but these are long out of print. These hardcover editions have become coveted collectors' items, and copies have sold for over [[US dollar|US$]]1000 on [[eBay]].{{Fact|date=September 2007}} |
|||
=[[Protein biosynthesis]]= |
|||
A [[Protein|protein]] is a [[Organic|naturally occurring]] [[Aliphatic|aliphatic]] [[Compound (chemistry)|compound]], composed of [[Amino acid|amino acid]]s (aa) and containing primarily at least the elements [[Carbon|carbon]] (C), [[Hydrogen|hydrogen]] (H), [[Nitrogen|nitrogen]] (N), and [[Oxygen|oxygen]] (O). Many proteins also contain [[Sulfur|sulfur]] (S), occasionally [[Phosphorus|phosphorus]] (P), and some [[Iron|iron]] (Fe). |
|||
Amino acids combine in a [[Condensation reaction|condensation reaction]] that releases water ([[Dehydration reaction|dehydration reaction]]) to form a [[Residue (chemistry)|residue]] held together by a [[Peptide bond|peptide bond]]. The [[Digestion|digestion]] of [[Protein in nutrition|dietary protein]]s produces [[Dipeptide|dipeptide]]s which are [[Absorption (chemistry)|absorbed]] more rapidly than aa. Some [[Tripeptide|tripeptide]]s and [[Tetrapeptide|tetrapeptide]]s are synthesized in humans. Proteins are [[Peptide|polypeptide molecules]]. Each protein is defined by a unique sequence of amino acid residues. This sequence is the [[Primary structure|primary structure]] of the protein. |
|||
In [[Biosynthesis|biosynthesis]], [[Precursor|precursor]]s are [[Activation energy|activated]] often with [[Nucleotide|nucleotide]]s, generally in the presence of a [[Catalysis|catalyst]], [[Redox|oxidizing or reducing agent]], to interconvert [[Biochemistry|biochemicals]] within living [[Organism|organism]]s. |
|||
=[[Amino acid synthesis]]= |
|||
[[Metabolic pathways]]) produce [[amino acid]]s from other compounds. The [[Substrate|substrate]]s for these pathways are usually [[Ingestion|ingested]] or synthesized internally by [[Gut flora|intestinal microbiota]] or activation of the [[Heterochromatin|heterochromatin]]. Of the [[Proteinogenic amino acid|proteinogenic]] aa, humans are able to synthesise up to 12 at the level needed for normal growth. The remaining [[Essential amino acid|essential]] aa must be supplied with or from food.<ref>{{cite journal |author=Young VR |title=Adult amino acid requirements: The case for a major revision in current recommendations |journal=J. Nutr. |volume=124 |issue=8 Suppl |pages=1517S–1523S |year=1994 |pmid=8064412 |url=http://jn.nutrition.org/cgi/reprint/124/8_Suppl/1517S.pdf}}</ref> |
|||
{| class="wikitable" |
|||
! Essential |
|||
! Nonessential |
|||
|- |
|||
| [[Isoleucine]] |
|||
| [[Alanine]] |
|||
|- |
|||
| [[Leucine]] |
|||
| [[Asparagine]] |
|||
|- |
|||
| [[Lysine]] |
|||
| [[Aspartate]] |
|||
|- |
|||
| [[Methionine]] |
|||
| [[Cysteine]]* |
|||
|- |
|||
| [[Phenylalanine]] |
|||
| [[Glutamate]] |
|||
|- |
|||
| [[Threonine]] |
|||
| [[Glutamine]]* |
|||
|- |
|||
| [[Tryptophan]] |
|||
| [[Glycine]]* |
|||
|- |
|||
| [[Valine]] |
|||
| [[Proline]]* |
|||
|- |
|||
| |
|||
| [[Serine]]* |
|||
|- |
|||
| |
|||
| [[Tyrosine]]* |
|||
|- |
|||
| |
|||
| [[Arginine]]* |
|||
|- |
|||
| |
|||
| [[Histidine]]* |
|||
|} |
|||
(*) Essential only in certain cases.<ref>{{cite journal |author=Fürst P, Stehle P |title=What are the essential elements needed for the determination of amino acid requirements in humans? |journal=J. Nutr. |volume=134 |issue=6 Suppl |pages=1558S–1565S |year=2004 |pmid=15173430 |url=http://jn.nutrition.org/cgi/content/full/134/6/1558S}}</ref><ref>{{cite journal |author=Reeds PJ |title=Dispensable and indispensable amino acids for humans |journal=J. Nutr. |volume=130 |issue=7 |pages=1835S–40S |year=2000 |pmid=10867060 |url=http://jn.nutrition.org/cgi/content/full/130/7/1835S}}</ref> |
|||
The situation is a more complicated since [[cysteine]], [[tyrosine]], [[histidine]] and [[arginine]] are semiessential aa in children. Their metabolic pathways are not fully developed.<ref>{{cite journal |author=Imura K, Okada A |title=Amino acid metabolism in pediatric patients |journal=Nutrition |volume=14 |issue=1 |pages=143–8 |year=1998 |pmid=9437700 |url=http://jn.nutrition.org/cgi/content/full/130/7/1835S |doi=10.1016/S0899-9007(97)00230-X}}</ref> The aa amounts required depend on the age and health of the individual. |
|||
In the [[citric acid cycle]] glutamate dehydrogenase catalyzes the reductive [[amination]] of α-ketoglutarate to glutamate: |
|||
<math>\alpha -ketoglutarate + NH_4^+ \rightleftarrows Glutamate</math> |
|||
At this step, the chirality of the amino acid is established. Afterwards, [[alanine]] and [[aspartate]] are synthesized by the [[transamination]] of [[glutamate]], vis a vis the transamination of [[pyruvate]] and [[oxaloacetate]], respectively. All of the remaining amino acids are constructed from [[glutamate]] or [[aspartate]], by [[transamination]] of these two amino acids with one α-keto acid. Glutamine is synthesized from NH<sub>4</sub><sup>+</sup> and glutamate, and [[asparagine]] is synthesized similarly. [[Proline]] and [[arginine]] are derived from glutamate. [[Serine]] formed from 3-phosphoglycerate, is the precursor of [[glycine]] and [[cysteine]]. [[Tyrosine]] is synthesized by the hydroxylation of [[phenylalanine]], an essential amino acid. |
|||
Most of the pathways of amino acid biosynthesis are regulated by [[feedback inhibition]], in which the committed step is allosterically inhibited by the final product. Branched pathways require extensive interaction among the branches that includes both negative and positive regulation. The regulation of glutamine synthetase from [[E. coli]] is a striking demonstration of cumulative feedback inhibition and of control by a cascade of reversible covalent modifications. |
|||
=Nucleotide synthesis= |
|||
==De novo synthesis== |
|||
===Purine nucleotides=== |
|||
[[Image:Nucleotides syn1.png|thumb|right|300px|<div style="border-width: 0px; border-bottom: 1px solid black; text-align: left;">'''The synthesis of IMP'''.</div>The color scheme is as follows: <span style="font-weight: bold;"><span style="color: blue;">enzymes</span>, <span style="color: rgb(219,155,36);">coenzymes</span>, <span style="color: rgb(151,149,45);">substrate names</span>, <span style="color: rgb(227,13,196);">metal ions</span>, <span style="color: rgb(128,0,0);">inorganic molecules</span> </span>]] |
|||
===Pyrimidine nucleotides=== |
|||
[[Image:Nucleotides_syn2.png|thumb|center|400px|<div style="border-width: 0px; border-bottom: 1px solid black; text-align: left;">'''The synthesis of UMP'''.</div>The color scheme is as follows: <span style="font-weight: bold;"><span style="color: blue;">enzymes</span>, <span style="color: rgb(219,155,36);">coenzymes</span>, <span style="color: rgb(151,149,45);">substrate names</span>, <span style="color: rgb(128,0,0);">inorganic molecules</span> </span>]] |
|||
==Salvage synthesis== |
|||
==[[Adenosine triphosphate]] (ATP)== |
|||
===[[Anaerobic respiration]]=== |
|||
[[Glucose]] C<sub>6</sub>H<sub>12</sub>O<sub>6</sub> <math>\to</math> 2 C<sub>3</sub>H<sub>6</sub>O<sub>3</sub> [[Lactic acid]] + 2 ATP. |
|||
===De novo synthesis of [[Purine|purine]] [[Nucleotide|nucleotide]]s=== |
|||
===Salvage synthesis=== |
|||
# [[Adenine]] + [[Phosphoribosyl pyrophosphate]] PRPP <----> [[Adenosine monophosphate|AMP]] + PP<sub>i</sub> : Enzyme ([[Adenine phosphoribosyltransferase]]) APRT<ref name="entrez">{{cite web | title = Entrez Gene: APRT adenine phosphoribosyltransferase| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=353| accessdate = }}</ref>, GeneID: 353. |
|||
==[[Cytidine triphosphate]] (CTP)== |
|||
Uridine 5'-triphosphate (UTP) + Glutamine + ATP + H<sub>2</sub>O => [[Cytidine triphosphate|Cytidine 5'-triphosphate]] + [[Glutamic acid|Glutamate]] + ADP + Orthophosphate (P<sub>i</sub>) : Enzyme ([[CTP synthase|Cytidine 5' triphosphate synthetase]]) CTPS, GeneID: 1503. |
|||
==[[Guanosine triphosphate]] (GTP)== |
|||
==[[Uridine triphosphate]] (UTP)== |
|||
De novo synthesis of the pyrimidine ring and conversion to UMP: |
|||
# L-[[Glutamine]] (Gln) + 2 ATP + HCO<sub>3</sub><sup>-</sup> + H<sub>2</sub>O => [[Carbamoyl phosphate]] + 2 ADP + Orthophosphate (P<sub>i</sub>) + [[Glutamic acid]] (Glu) : Enzyme 1 (Carbomoyl-phosphate Synthetase 2, CPS II) CAD(E1), GeneID: 790. |
|||
# Carbamoyl phosphate + L-[[Aspartic acid|Aspartate]] <=> N-carbamoyl L-aspartate + Orthophosphate (P<sub>i</sub>) : Enzyme 2 (Aspartate transcarbamylase, ATCase) CAD(E2), GeneID: 790. |
|||
# N-Carbamoyl L-Aspartate + H+ <=> (S)-Dihydroorotate + H<sub>2</sub>O : Enzyme 3 ([[Dihydroorotase]], DHO) CAD(E3), GeneID: 790. |
|||
# (S)-Dihydroorotate + [[Coenzyme Q10|Ubiquinone]] => [[Orotic acid|Orotate]] + [[Coenzyme Q10|Ubiquinol]] : Enzyme ([[Dihydroorotate dehydrogenase]]) DHODH, GeneID: 1723 |
|||
# Orotate + [[Phosphoribosyl pyrophosphate|5-Phospho-alpha-D-Ribose 1-Diphosphate]] (PRPP) <=> [[Orotidine 5'-monophosphate]] (OMP) + [[Pyrophosphate|PP<sub>i</sub>]] : Enzyme 1 ([[Uridine monophosphate synthetase]]) UMPS(E1), GeneID: 7372. |
|||
# Orotidine 5'-monophosphate => [[Uridine monophosphate|Uridine 5'-monophosphate]] (UMP) + CO<sub>2</sub> : Enzyme 2 UMPS(E2), GeneID: 7372 |
|||
# Uridine 5'-monophosphate (UMP) + ATP <=> [[Uridine diphosphate|Uridine 5'-diphosphate]] (UDP) + ADP : Enzymes (Uridine monophosphate kinase) UMPK, GeneID: 51727; (uridine-cytidine kinase 2) [[UCK2]], GeneID:7371. |
|||
# Uridine 5'-diphosphate (UDP) + ADP <=> [[Uridine triphosphate|uridine 5'-triphosphate]] (UTP) + AMP : Enzymes ([[Nucleoside-diphosphate kinase]]s A, B, C, D; NME6, NME7) NDKA-D, GeneIDs: 4830, 4831, 4832, 4833; NME6 GeneID: 10201, NME7 GeneID: 29922. |
|||
=[[Transcription (genetics)|Transcription]]= |
|||
A [[DNA]] sequence is [[Enzyme|enzymatically]] copied by [[RNA polymerase II]] to produce a complementary nucleotide [[RNA]] strand of messenger RNA (mRNA). mRNA is modified through [[RNA splicing]], 5' end capping, and the addition of a polyA tail.<ref> Robert J. Brooker ''Genetics: analysis and principles.'' 2nd edition. (New York: McGraw-Hill 2005) Chapter 12 "Gene transcription and RNA modification" pp. 318-325.</ref> |
|||
===[[Transcriber]]=== |
|||
In human [[Biochemistry|biochemistry]] a transcriber might be thought of as a tool assembled in the nucleus for the transcription of genetic information. It synthesizes (a nucleic acid, usually RNA) using an existing nucleic acid (DNA) as a template from which to copy. |
|||
===[[Transcription factor|Transcription factors]]=== |
|||
[[Transcription factor|Transcription factors]] bind to specific [[DNA sequence|sequences]] of [[DNA]] and are part of the system that controls the [[transcription (genetics)|transcription]] of genetic information from DNA to [[RNA]].<ref name="pmid9570129">{{cite journal | author = Latchman DS | title = Transcription factors: an overview | journal = Int. J. Biochem. Cell Biol. | volume = 29 | issue = 12 | pages = 1305–12 | year = 1997 | pmid = 9570129 | doi = 10.1016/S1357-2725(97)00085-X }}</ref><ref name="pmid2128034">{{cite journal | author = Karin M | title = Too many transcription factors: positive and negative interactions | journal = New Biol. | volume = 2 | issue = 2 | pages = 126–31 | year = 1990 | pmid = 2128034 | doi = | issn = }}</ref> Transcription factors occur in a complex, increase (as an [[Activator (genetics)|activator]]), or prevent (as a [[repressor]]) the presence of [[RNA polymerase II]].<ref name="pmid8870495">{{cite journal | author = Roeder RG | title = The role of general initiation factors in transcription by RNA polymerase II | journal = Trends Biochem. Sci. | volume = 21 | issue = 9 | pages = 327–35 | year = 1996 | pmid = 8870495 | doi = 10.1016/0968-0004(96)10050-5 }}</ref><ref name="pmid8990153">{{cite journal | author = Nikolov DB, Burley SK | title = RNA polymerase II transcription initiation: a structural view | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 94 | issue = 1 | pages = 15–22 | year = 1997 | pmid = 8990153 | doi = 10.1073/pnas.94.1.15 }}</ref><ref name="pmid11092823">{{cite journal | author = Lee TI, Young RA | title = Transcription of eukaryotic protein-coding genes | journal = Annu. Rev. Genet. | volume = 34 | issue = | pages = 77–137 | year = 2000 | pmid = 11092823 | doi = 10.1146/annurev.genet.34.1.77}}</ref> |
|||
Each transcription factor contains one or more [[DNA binding domain]]s (DBDs) which attach to specific sequences of DNA adjacent to the gene that it regulates.<ref name="pmid2667136">{{cite journal | author = Mitchell PJ, Tjian R | title = Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins | journal = Science | volume = 245 | issue = 4916 | pages = 371-8 | year = 1989 | pmid = 2667136 | doi = 10.1126/science.2667136 | issn = }}</ref><ref name="pmid9121580">{{cite journal | author = Ptashne M, Gann A | title = Transcriptional activation by recruitment | journal = Nature | volume = 386 | issue = 6625 | pages = 569-77 | year = 1997 | pmid = 9121580 | doi = 10.1038/386569a0 | issn = }}</ref> |
|||
There are approximately 2600 proteins in the [[human genome]] that contain DNA-binding domains and most of these are presumed to function as transcription factors.<ref name="pmid15193307">{{cite journal | author = Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA | title = Structure and evolution of transcriptional regulatory networks | journal = Curr. Opin. Struct. Biol. | volume = 14 | issue = 3 | pages = 283–91 | year = 2004 | pmid = 15193307 | doi = 10.1016/j.sbi.2004.05.004 }}</ref> |
|||
The combinatorial use of a subset of the approximately 2000 human transcription factors easily accounts for the unique regulation of each gene in the human genome during [[developmental biology|development]].<ref name="pmid11823631" /> |
|||
[[General transcription factor]]s (GTFs) are necessary for transcription to occur. GTFs are part of the large transcription [[preinitiation complex]] that interacts with [[RNA polymerase II]] directly.<ref name="pmid16858867">{{cite journal | author = Thomas MC, Chiang CM | title = The general transcription machinery and general cofactors | journal = Critical reviews in biochemistry and molecular biology | volume = 41 | issue = 3 | pages = 105–78 | year = 2006 | pmid = 16858867 | doi = | url = | issn = }}</ref> They are ubiquitous and interact with the core promoter region surrounding the transcription start site(s) of all[[class II gene]]s.<ref name="pmidc">{{cite journal | author = Orphanides G, Lagrange T, Reinberg D | title = The general transcription factors of RNA polymerase II | journal = Genes Dev. | volume = 10 | issue = 21 |pages = 2657–83 | year = 1996 | pmid = 8946909 | doi = 10.1101/gad.10.21.2657 }}</ref> |
|||
[[TFIIA]] |
|||
[[TFIIB]] |
|||
[[TFIID]] (see also [[TATA binding protein]]) |
|||
[[TFIIE]] |
|||
[[TFIIF]] |
|||
[[TFIIH]] |
|||
Active transcription units (transcription factors) are clustered in the nucleus. There are ~10,000 factors in the nucleoplasm of a [[HeLa cell]], among which are ~8,000 polymerase II factors and ~2,000 polymerase III factors. Each polymerase II factor contains ~8 polymerases. As most active transcription units are associated with only one polymerase, each factor will be associated with ~8 different transcription units. These units might be associated through promoters and/or enhancers, with loops forming a ‘cloud’ around the factor. |
|||
Additional proteins: |
|||
[[coactivator (genetics)|coactivator]]s |
|||
[[Chromatin Structure Remodeling (RSC) Complex|chromatin remodeler]]s |
|||
[[histone acetyltransferase|histone acetylases]] |
|||
[[histone deacetylase| deacetylases]] |
|||
[[kinase]]s |
|||
[[methylase]]s |
|||
play crucial roles in [[regulation of gene expression|gene regulation]].<ref name="pmid11823631">{{cite journal | author = Brivanlou AH, Darnell JE | title = Signal transduction and the control of gene expression | journal = Science | volume = 295 | issue = 5556 | pages = 813-8 | year = 2002 | pmid = 11823631 | doi = 10.1126/science.1066355 | issn = }}</ref> |
|||
The '''TATA binding protein''' ('''TBP''', GeneID: 6908) is a [[transcription factor]] that binds specifically to a [[DNA]] sequence called the [[TATA box]]. This DNA sequence is found about 25-30 [[base pair]]s upstream of the transcription start site in some [[eukaryote|eukaryotic]] [[gene]] [[promoter]]s.<ref name="pmid17670940">{{cite journal | author = Kornberg RD | title = The molecular basis of eukaryotic transcription | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 104 | issue = 32 | pages = 12955–61 | year = 2007 | pmid = 17670940 | doi = 10.1073/pnas.0704138104}}</ref> TBP, along with a variety of [[TBP-associated factor]]s, make up the [[TFIID]], a [[general transcription factor]] that in turn makes up part of the [[RNA polymerase II]] [[preinitiation complex]].<ref name="pmid11092823">{{cite journal | author = Lee TI, Young RA | title = Transcription of eukaryotic protein-coding genes | journal = Annu. Rev. Genet. | volume = 34 | issue = | pages = 77–137 | year = 2000 | pmid = 11092823 | doi = 10.1146/annurev.genet.34.1.77}}</ref> As one of the few proteins in the preinitation complex that binds DNA in a sequence-specific manner, it helps position RNA polymerase II over the transcription start site of the gene. However, it is estimated that only 10-20% of human promoters have TATA boxes. Therefore, TBP is probably not the only protein involved in positioning RNA polymerase II. |
|||
TBP is a subunit of the eukaryotic [[transcription factor]] TFIID. [[TFIID]] is the first protein to bind to DNA during the formation of the pre-initiation [[Transcription (genetics)|transcription]] complex of [[RNA polymerase II]] (RNA Pol II). Binding of TFIID to the [[TATA box]] in the [[promoter]] region of the gene initiates the recruitment of other factors required for RNA Pol II to begin transcription. Some of the other recruited [[transcription factors]] include [[TFIIA]], [[TFIIB]] and [[TFIIF]]. Each of these transcription factors are formed from the interaction of many protein subunits, indicating that transcription is a heavily regulated process. |
|||
When TBP binds to a [[TATA box]] within the [[DNA]], it distorts the DNA by inserting amino acid side chains between base pairs, partially unwinding the helix, and doubly kinking it. The distortion is accomplished through a great amount of surface contact between the protein and DNA. TBP binds with the negatively charged phosphates in the DNA backbone through positively charged [[lysine]] and [[arginine]] amino acid residues. The sharp bend in the DNA is produced through projection of four bulky [[phenylalanine]] residues into the minor groove. As the DNA bends, its contact with TBP increases, thus enhancing the DNA-protein interaction. |
|||
The strain imposed on the DNA through this interaction initiates melting, or separation, of the strands. Because this region of DNA is rich in [[adenine]] and [[thymine]] residues, which base pair through only two [[hydrogen bonds]], the DNA strands are more easily separated. Separation of the two strands exposes the bases and allows [[RNA polymerase II]] to begin transcription of the [[gene]]. |
|||
===[[RNA polymerase II]]=== |
|||
Each of the [[RNA polymerase|RNA polymerases]] transcribes different classes of proteins. Specifically, [[RNA polymerase I]] transcribes [[rRNA]] and [[RNA polymerase III]] transcribes various additional RNAs. |
|||
[[RNA polymerase II]] transcribes [[Class II gene|Class II genes]] to form proteins. It catalyzes the [[Transcription (genetics)|transcription]] of [[DNA]] to synthesize precursors of [[mRNA]] and most [[snRNA]] and [[microRNA]].<ref>{{cite journal | author = Kronberg, R.D. | title = Eukaryotic transcriptional control | journal = Trends Cell Biol. | year=1999 | volume=9 | issue=12 | pages=M46–49 | pmid = 10611681 | doi = 10.1016/S0962-8924(99)01679-7}}</ref> |
|||
It is a 550 [[kDa]] complex of 12 subunits: |
|||
'''Polymerase (RNA) II (DNA directed) polypeptide A, 220kDa''' ([[POLR2A]]), GeneID: 5430. |
|||
This largest subunit contains a carboxy terminal domain composed of heptapeptide repeats that are essential for polymerase activity. These repeats contain serine and threonine residues that are phosphorylated in actively transcribing RNA polymerase. In addition, this subunit, in combination with several other polymerase subunits, forms the DNA binding domain of the polymerase, a groove in which the DNA template is transcribed into RNA.<ref>{{cite web | title = Entrez Gene: POLR2A polymerase (RNA) II (DNA directed) polypeptide A, 220kDa| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5430| accessdate = }}</ref> |
|||
'''Polymerase (RNA) II (DNA directed) polypeptide B, 140kDa''' ([[POLR2B]]), GeneID: 5431. |
|||
This second largest subunit, in combination with at least two other polymerase subunits, forms a structure within the polymerase that maintains contact in the active site of the enzyme between the DNA template and the newly synthesized RNA.<ref>{{cite web | title = Entrez Gene: POLR2B polymerase (RNA) II (DNA directed) polypeptide B, 140kDa| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5431| accessdate = }}</ref> |
|||
'''Polymerase (RNA) II (DNA directed) polypeptide C, 33kDa''' ([[POLR2C]]), GeneID: 5432. |
|||
This third largest subunit contains a cysteine rich region and exists as a heterodimer with another polymerase subunit, POLR2J. These two subunits form a core subassembly unit of the polymerase.<ref>{{cite web | title = Entrez Gene: POLR2C polymerase (RNA) II (DNA directed) polypeptide C, 33kDa| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5432| accessdate = }}</ref> |
|||
'''DNA-directed RNA polymerase II polypeptide D (or subunit B4)''' ([[POLR2D]]), GeneID: 5433. |
|||
This is the fourth largest subunit.<ref>{{cite web | title = Entrez Gene: POLR2D polymerase (RNA) II (DNA directed) polypeptide D| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5433| accessdate = }}</ref> |
|||
'''Polymerase (RNA) II (DNA directed) polypeptide E, 25kDa''' ([[POLR2E]]), GeneID: 5434. |
|||
This the fifth largest subunit is present in two-fold molar excess over the other polymerase subunits. An interaction may occur between transcriptional activators and the polymerase through this subunit.<ref>{{cite web | title = Entrez Gene: POLR2E polymerase (RNA) II (DNA directed) polypeptide E, 25kDa| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5434| accessdate = }}</ref> |
|||
'''Polymerase (RNA) II (DNA directed) polypeptide F''' ([[POLR2F]]), GeneID: 5435. |
|||
This sixth largest subunit, based on studies in yeast, in combination with at least two other subunits, may form a structure that stabilizes the transcribing polymerase on the DNA template.<ref>{{cite web | title = Entrez Gene: POLR2F polymerase (RNA) II (DNA directed) polypeptide F| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5435| accessdate = }}</ref> |
|||
'''Polymerase (RNA) II (DNA directed) polypeptide G''' ([[POLR2G]]), GeneID: 5436. |
|||
==Books of the Satanic Bible== |
|||
{{Original research|date=September 2007}} |
|||
This seventh largest subunit, from studies in yeast, may play a role in regulating polymerase function.<ref>{{cite web | title = Entrez Gene: POLR2G polymerase (RNA) II (DNA directed) polypeptide G| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5436| accessdate = }}</ref> |
|||
===Dedication=== |
|||
'''Polymerase (RNA) II (DNA directed) polypeptide H''' ([[POLR2H]]), GeneID: 5437. |
|||
Long since removed from contemporary printings of the book, the first edition of The Satanic Bible contained an extensive dedication to the thinkers who influenced LaVey. |
|||
This is an essential subunit.<ref>{{cite web | title = Entrez Gene: POLR2H polymerase (RNA) II (DNA directed) polypeptide H| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5437| accessdate = }}</ref> |
|||
The primary dedication of the book was made to [[Bernadino Nogara]] (misprinted as "Logara"), |
|||
[[Karl Haushofer]], [[Rasputin]], [[Basil Zaharoff|Sir Basil Zaharoff]], [[Alessandro Cagliostro]], |
|||
Barnabas Saul (Dr. John Dee's first Scryer), [[Ragnar Redbeard]], [[William Mortensen]], Hans Brick, [[Max Reinhardt (theatre director)|Max Reinhardt]], the American Sociologist Orrin Klapp, [[Fritz Lang]], [[Friedrich Nietzsche]], [[W. C. Fields]], [[P. T. Barnum]], [[Hans Poelzig]], [[Reginald Marsh (artist)|Reginald Marsh]], [[Wilhelm Reich]] and [[Mark Twain]]. |
|||
'''Polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa''' ([[POLR2I]]), GeneID: 5438. |
|||
The secondary dedication included [[Howard Hughes]], [[Marcello Truzzi]], [[Marilyn Monroe]], |
|||
[[William Lindsay Gresham]], Hugo Zacchini,<ref>http://www.cannon-mania.com/human-cannon.htm Hugo Zacchini</ref> [[Jayne Mansfield]], Fredrick Goerner, [[Nathaniel West]], [[Horatio Alger]], |
|||
[[Robert E. Howard]], [[George Orwell]], [[H. P. Lovecraft]], [[Tuesday Weld]], |
|||
[[H.G. Wells]], [[Houdini|Harry Houdini]], Togare (LaVey's pet lion) and [[Nine Unknown Men|The Nine Unknown Men]]. |
|||
This subunit in combination with two other polymerase subunits forms the DNA binding domain of the polymerase, a groove in which the DNA template is transcribed into RNA. The product of this gene has two zinc finger motifs with conserved cysteines and the subunit does possess zinc binding activity.<ref>{{cite web | title = Entrez Gene: POLR2I polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5438| accessdate = }}</ref> |
|||
''The Satanic Bible'', after the introductions by other authors, is divided into four books: the ''Book of [[Satan]]'', the ''Book of [[Lucifer]]'', the ''Book of [[Belial]]'', and finally the ''Book of [[Leviathan]]''. LaVey seems to have taken this hierarchy from [[The Book of the Sacred Magic of Abramelin the Mage]], in which these four demons serve as the chiefs of Hell. Each book approaches a different aspect of Satanism, and serves a unique purpose within the structure of ''The Satanic Bible''. |
|||
'''Polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa''' ([[POLR2J]]), GeneID: 5439. |
|||
===The Book of Satan=== |
|||
''The Book of Satan: The Infernal Diatribe'' introduces Satanism in dramatic fashion: through the verse of [[Ragnar Redbeard]] in [[Might is Right]][http://www.plausiblefutures.com/getfile.php/51711.476/might.pdf ]. Anton LaVey chose segments from different sections of ''Might is Right'', and made slight changes to the verse to amend what he perceived as errors in logic or consistency with ''Might is Right''. As Anton LaVey stated in his introduction to later editions of ''Might is Right'', he found both considerable inspiration in that book, but also glaring errors that made it, taken part by part, incompatible with his Satanic philosophy; it was instead the whole of the message that he found appealing, and the passages selected and changes made to best capture what he found appealing in ''Might is Right''. ''The Book of Satan'' was also recited by LaVey in ritual ceremonies, in part or in whole, and his recording of ''The Satanic Mass'' includes a full recitation of ''The Book of Satan''. |
|||
This subunit exists as a heterodimer with another polymerase subunit; forming a core subassembly unit of the polymerase. Two similar genes are located nearby on chromosome 7q22.1.<ref>{{cite web | title = Entrez Gene: POLR2J polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5439| accessdate = }}</ref> |
|||
''The Book of Satan'' is symbolically associated with the element of [[Fire]]. |
|||
'''DNA directed RNA polymerase II polypeptide J-related gene''' ([[POLR2J2]]), GeneID: 246721.<ref>{{cite web | title = Entrez Gene: POLR2J2 DNA directed RNA polymerase II polypeptide J-related gene| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=246721| accessdate = }}</ref> |
|||
===The Book of Lucifer: The Enlightenment=== |
|||
''The Book of Lucifer: The Enlightenment'' directly follows ''The Book of Satan''. After the indignant intonation of ''The Book of Satan'', ''The Book of Lucifer'' seeks to logically expound the philosophy and dogma of Satanism. ''The Book of Lucifer'' is divided into twelve essays, each of them a vital component of LaVey's architecture of Satanism. The following are brief synopses of each of these essays: |
|||
This gene is a member of the RNA polymerase II subunit 11 gene family, which includes three genes in a cluster on chromosome 7q22.1. The founding member of this family, DNA directed RNA polymerase II polypeptide J, encodes multiple, alternatively spliced transcripts that potentially express isoforms with distinct C-termini compared to DNA directed RNA polymerase II polypeptide J. Most or all variants are spliced to include additional non-coding exons at the 3' end which makes them candidates for nonsense-mediated decay (NMD). Consequently, it is not known if this locus expresses a protein or proteins in vivo.<ref>{{cite web | title = Entrez Gene: POLR2J2 DNA directed RNA polymerase II polypeptide J-related gene| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=246721| accessdate = }}</ref> |
|||
'''I.''' - ''Wanted! - ''God - Dead or Alive'' |
|||
'''DNA directed RNA polymerase II polypeptide J3''' (POLR2J3), GeneID: 548644.<ref>{{cite web | title = Entrez Gene: POLR2J3 DNA directed RNA polymerase II polypeptide J3| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=548644| accessdate = }}</ref> |
|||
This short essay provides an essentially atheistic approach to Satanism, and indicts Judeo-Christian religion and prayer as hypocritical and unrealistic. Further, it clarifies the Satanic approach to the question of God: |
|||
''...the Satanist simply accepts the definition which suits him best. Man has always created his gods, rather than his gods creating him. To the Satanist "God"...is seen as the balancing force in nature, and not as being concerned with suffering.''<ref>LaVey, Anton ''The Satanic Bible'', pg 40</ref> |
|||
The passage from which this is taken, when viewed as a whole, suggests that "God" is a human concept, a means of explaining that which men have been unable to explain either through ignorance or through philosophical inability to grasp the nature of reality. This passage also opens up the common Satanic maxim, "I am my own god", a statement of self-aggrandizement, viewed as a positive trait by Satanists. |
|||
'''II.''' ''The God you Save May be Yourself'' |
|||
This gene is a member of the RNA polymerase II subunit 11 gene family, which includes three genes in a cluster on chromosome 7q22.1. This locus produces multiple, alternatively spliced transcripts that potentially express isoforms with distinct C-termini compared to DNA directed RNA polymerase II polypeptide J. Most or all variants are spliced to include additional non-coding exons at the 3' end which makes them candidates for nonsense-mediated decay (NMD). Consequently, it is not known if this locus expresses a protein or proteins in vivo.<ref>{{cite web | title = Entrez Gene: POLR2J2 DNA directed RNA polymerase II polypeptide J-related gene| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=548644| accessdate = }}</ref> |
|||
Follows up on the concept of "I am my own god" with a full explanation of the Satanic egocentric view of the world. This short essay states that as all gods are of human creation, worshipping an external god is to worship another human by proxy; therefore, the sensible, Satanic approach is to create your own god, namely yourself, and to "worship" this god. The result, of course, is to view oneself as the most important of all beings, and to adopt an unapologetically self-centered view of the world and course of action. |
|||
'''DNA directed RNA polymerase II polypeptide J4, pseudogene''' (POLR2J4), GeneID: 84820.<ref>{{cite web | title = Entrez Gene: POLR2J4 DNA directed RNA polymerase II polypeptide J4, pseudogene| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=84820| accessdate = }}</ref> |
|||
'''III.''' ''Some Evidence of the New Satanic Age'' |
|||
Although now thought to be a pseudogene, model predictions suggests that a miscRNA from this gene of possible transcription function may be involved in DNA binding and DNA-directed RNA polymerase activity.<ref>{{cite web | title = Entrez Gene: POLR2J2 DNA directed RNA polymerase II polypeptide J-related gene| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=84820| accessdate = }}</ref> |
|||
This essay is longer than the previous essays; following the first two that denounce traditional religion as hypocritical and self-hating, this one offers Satanism as an alternative and opposite, a religion suited to human needs. |
|||
'''Polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa''' ([[POLR2K]]), GeneID: 5540. |
|||
It begins by suggesting that the [[Seven Deadly Sins]] are in fact all instinctual to human nature and not sinful at all. It states that they are all unavoidable urges of mankind, carefully selected by Christianity to ensure that all men will inevitably sin, as no one can avoid engaging in these instinctive urges. LaVey submits that this is a device to guarantee that humans within the Christian religious framework will surely sin and have no choice but to beg God for forgiveness; therefore, dependence upon the Church is assured. Instead, LaVey states that as all of these so-called sins are natural to humans, they should be embraced and even considered virtuous. This excerpt, for example: |
|||
This is one of the smallest subunits.<ref>{{cite web | title = Entrez Gene: POLR2K polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5440| accessdate = }}</ref> |
|||
''Envy and greed are the motivating forces of ambition - and without ambition, very little of any importance would be accomplished.''<ref>LaVey, Anton ''The Satanic Bible'', pg 46</ref> |
|||
'''RP11-90M2.3 novel protein similar to Polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa''' (RP11-90M2.3), GeneID: 100129174. |
|||
He goes on to explain that in this modern age, religionists have had to constantly reinterpret their own texts in order to keep up with the demands of people that they be permitted to indulge their normal human desires. LaVey views this as both hypocritical and evidence that these religions are inherently obsolete and should be discarded entirely, to be replaced with a religion better suited to man's needs. Satanism, LaVey suggests, is that religion. |
|||
This maybe another small subunit.<ref>{{cite web | title = Entrez Gene: RP11-90M2.3 novel protein similar to polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa POLR2K| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=100129174| accessdate = }}</ref> |
|||
LaVey then justifies Satanism as a religion by explaining that it is not merely a philosophy. He explains that one reason man has always had religion is because he has a need for dogma and ceremony; Satanism acknowledges this, and therefore supplies its adherents with dogma and ceremony in the form of magic and ritual. LaVey claims that it is precisely this trait that distinguishes Satanism from [[Humanism]] or other essentially atheistic philosophies, and makes it a true religion suited to man's carnal nature. |
|||
'''Polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa''' ([[POLR2L]]), GeneID: 5441. |
|||
LaVey concludes the essay by explaining that even other religions or new-age movements that claim to supply adherents with magic have failed in this by distinguishing "white magic" from "black magic;" LaVey claims that all magic is one and the same, as all of it is done for the glorification of the magic user and therefore (like all human actions) is essentially selfish. He suggests that a Satanist may choose to help those he cares for, including himself, or condemn those he hates, but in all cases what he does is at his own discretion and therefore done for selfish reasons. |
|||
The subunit contains four conserved cysteines characteristic of an atypical zinc-binding domain.<ref>{{cite web | title = Entrez Gene: POLR2L polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5441| accessdate = }}</ref> |
|||
'''IV.''' ''Hell, the Devil, and How to Sell Your Soul'' |
|||
[[RNA polymerase II holoenzyme]] |
|||
In this essay LaVey now explains why, if he does not believe in literal concepts of gods or devils, he chose the name "Satanism" to describe his religion. |
|||
====Initiation Regulation==== |
|||
LaVey points out the Hebraic origin of the word "Satan" as a term to mean "adversary", not only applied to a supernatural being but to any person who is your opponent. He likewise describes the mythological and literary significance of [[Satan]] in history, from the Greek [[Pan (mythology)|Pan]] to the "scapegoat" used to absorb men's sins to the co-opting of pagan deities and devils by Christianity. |
|||
Initiation is regulated by several mechanisms: |
|||
LaVey's final assessment is that as Satan and all the devils have represented man's carnal nature made sinful, and the opposition of everything from servile god-worship to conformity, Satan as a literary and mythological figure is the ideal symbol for a religion that exalts man's carnal and independent nature. LaVey's view of Satan might be best compared to [[John Milton]]'s literary Satan, a proud and independent beast. |
|||
#Protein interference. |
|||
#[[Chromatin]] structure inhibition. |
|||
#Phosphorylation. |
|||
=====Regulation by Protein interference===== |
|||
'''V.''' ''Love and Hate'' |
|||
Protein interference is the process where some signaling protein interacts, either with the promoter or some stage of the partially constructed complex, to prevent further construction of the polymerase complex. This is generally a very rapid response and is used for fine level, individual gene control and for 'cascade' processes for a group of genes useful under a specific conditions (for example DNA repair genes or heat shock genes). |
|||
A surprisingly short essay given its importance, here LaVey explains in no uncertain terms how Satanists view matters of love and hate, and their role in human affairs. |
|||
=====Regulation by Chromatin structure inhibition===== |
|||
LaVey makes it very clear that although Satanism is an uncompromisingly selfish religion, he defines selfishness according to what an individual truly wants. Therefore, if a person should honestly care for another person and wishes to express love, then he should do so wholeheartedly; a truly selfish person can acknowledge that if a person is loved by him, then they are important by virtue of his love. This can be compared favorably to the arguments of [[ethical egoism]]—that what sometimes benefits others can be beneficial to oneself, but that one must always have one's own interests first in mind. LaVey never suggests that love is not a natural emotion in man, and on the contrary suggests that loving select individuals is very natural, but he does claim that to love all people is not only a philosophical mistake but is in fact impossible and even damaging to the ability to truly love those few individuals who deserve it. |
|||
[[Chromatin]] structure inhibition is the process where the promoter is hidden by [[chromatin]] structure. Chromatin structure is controlled by post-translational modification of the [[histone]]s involved and leads to gross levels of high or low transcription levels. See: [[chromatin]], [[histone]] and [[nucleosome]]. |
|||
LaVey explains that hatred is likewise a natural emotion in man and therefore not to be shunned. He makes clear that hatred should be directed at those who deserve it by virtue of their actions to offend the individual, and like love, it is senseless to universally apply hatred to all mankind. He muses that while Satanism strongly advocates both individual love and hate, because white-light religion has such a strong aversion to acknowledging hate as a natural feeling in man that to merely mention that Satanism permits individuals to hate their enemies, Satanism is automatically portrayed as a hateful religion, a claim he maintains is false and ignorant of the true ethics of Satanism. |
|||
These methods of control can be combined in a modular method, allowing very high specificity in transcription initiation control. |
|||
'''VI.''' ''Satanic Sex'' |
|||
=====Regulation by Phosphorylation===== |
|||
Contrary to the popular opinion that Satanism advocates promiscuous behavior in all individuals, in this essay LaVey actually lambasts the "free love" movement (a movement very much in motion in the 1960s when LaVey wrote ''The Satanic Bible'') as being equally restricting as the white-light view that any unholy sex is wrong. |
|||
POLR2A has a domain at its C-terminus, the CTD (C-terminal domain) which is the target of [[protein kinase|kinases]] and [[phosphatases]]. The phosphorylation of the CTD is an important regulation mechanism, as this allows attraction and rejection of factors that have a function in the transcription process. The CTD can be considered as a platform for [[transcription factors]]. |
|||
LaVey's stance, once again, takes a purely individual approach to sexual matters and ethics. He maintains that while some people are indeed happy with sexual promiscuity, some are, by their nature, happier with much less sexual activity, or perhaps no sex activity at all. LaVey believes that neither of these states is unnatural or deserving of condemnation, but rather that it is a decision for each individual to make concerning their own sexual tastes and activities. |
|||
The CTD consists of repetitions of an [[amino acid]] motif, YSPTSPS, of which [[Serine]]s and [[Threonine]]s can be [[phosphorylated]]. The mammalian protein contains 52. There are many different combinations of phosphorylations possible on these repeats and these can change rapidly during transcription. The regulation of these phosphorylations and the consequences for the association of transcription factors plays a major role in the regulation of transcription. |
|||
From this basic principle, LaVey then expounds upon this by pointing out exactly what is and is not permissible Satanic sexual activity. |
|||
During the transcription cycle, the CTD of POLR2A is reversibly phosphorylated. RNAP II containing unphosphorylated CTD is recruited to the promoter, whereas the hyperphosphorylated CTD form is involved in active transcription. Phosphorylation occurs at two sites within the heptapeptide repeat, at Serine 5 and Serine 2. Serine 5 phosphorylation is confined to promoter regions and is necessary for the initiation of transcription, whereas Serine 2 phosphorylation is important for mRNA elongation and 3'-end processing. |
|||
The basic premise of what is permissible is summed up by the maxim: |
|||
===[[Mediator (coactivator)]]=== |
|||
''Satanism encourages any form of sexual expression you may desire, so long as it hurts no one else.''<ref>LaVey, Anton ''The Satanic Bible'', pg 69</ref> |
|||
==Messenger RNA== |
|||
LaVey quickly explains that this does not preclude sexual sadism/masochism, as "so long as it hurts no one else" must be interpreted to mean "who does not wish to be hurt." |
|||
[[Image:MRNA-interaction.png|thumb|300px| |
|||
This statement openly condones [[homosexuality]], [[bisexuality]], [[polyamory]], premarital or extramarital sex, sexual games including [[BDSM]], multiple partners, [[incest]], and any other such proclivity, while at the same time not excluding [[heterosexuality]], [[monogamy]], or "traditional" marriage. Satanism views all such activities as entirely equal,{{Fact|date=February 2007}} and deserving of the same respect. LaVey also specifies asexuality as a valid expression, for one for whom sexual activity is simply not desired. LaVey claims Satanism to be the first religion to openly take this stance. |
|||
The "life cycle" of an '''mRNA''' in a eukaryotic cell. RNA is [[transcription (genetics)|transcribed]] in the [[cell nucleus|nucleus]]; once completely processed, it is transported to the [[cytoplasm]] and [[Translation (genetics)|translated]] by the [[ribosome]]. At the end of its life, the mRNA is degraded.]] |
|||
In the ribosome, the nucleic acid polymer is [[translation (genetics)|translated]] into a polymer of [[amino acids]]: a protein. |
|||
[[Image:Transcription label fromcommons.jpg|thumb|250px|right|A micrograph of ongoing gene transcription of [[ribosomal RNA]] illustrating the growing [[primary transcript]]s. "Begin" indicates the [[3' end]] of the DNA template strand, where new RNA synthesis begins; "end" indicates the [[5' end]], where the primary transcripts are almost complete.]] |
|||
However, the same statement therefore excludes any such activity as [[rape]], [[pedophilia]], or other sexual activities in which any of the participants are unwilling or unable to give knowledgeable consent{{Fact|date=October 2007}} (as is the case with a child or animal). Satanism also expressly forbids illegal activity of any kind.{{Fact|date=October 2007}} |
|||
==Pre-Initiation== |
|||
'''VII.''' ''Not All Vampires Suck Blood'' |
|||
RNA polymerase II binds to the DNA and, along with other cofactors, unwinds the DNA to create an initiation bubble so that the polymerase has access to the single-stranded DNA template. RNA Polymerase II does require a promoter like sequence. |
|||
One of the most famous essays from ''The Satanic Bible'', it is here that LaVey coins the term "psychic vampire". LaVey defines a psychic vampire as one who attempts to psychologically manipulate others by systematically playing the victim; for example, a person who constantly uses some minor physical flaw as an excuse for their shortcomings and a means of gaining sympathy and favor from others. LaVey believes that people who use a victim status as a means to induce guilt in others are fundamentally weak, and therefore to be shunned by Satanists. (LaVey does not imply that anyone with a flaw is automatically weak, but rather that the use of that flaw to gain sympathy and favor is weak.) |
|||
Proximal (core) Promoters: |
|||
LaVey advises that such people are psychologically draining (hence the term "psychic vampire") and should be dealt with mercilessly and discarded before they are permitted to take control of the lives of vital individuals. |
|||
TATA promoters are found around -30 bp to the start site of transcription. Not all genes have TATA box promoters and there exists TATA-less promoters as well. The TATA promoter consensus sequence is TATA(A/T)A(A/T). |
|||
The following are the steps involved in TATA Promoter Complex formation: |
|||
'''VIII.''' ''Indulgence NOT Compulsion'' |
|||
1. General transcription factors bind |
|||
Here, LaVey puts forth Satanism as an essentially hedonistic philosophy; however, LaVey's approach to hedonism is [[epicureanism|epicurean]] in nature. |
|||
2. TFIID, TFIIA, TFIIB, TFIIF (w/RNA Polymerase II), TFIIH/E |
|||
LaVey states clearly that he believes that man should always tend towards indulgence, not abstinence. Whereas other religions seek to dictate what man should abstain from (which according to LaVey, is most of man's natural urges), Satanism seeks to encourage man to indulge all his carnal desires (so long as they fall within the bounds of Satanic ethics, see the essay on Satanic sexuality for example). Satanism, as an atheistic religion, holds that as there is no afterlife and therefore no paradise or [[heaven]] or [[hell]], all happiness and satisfaction must be attained here, on earth. LaVey therefore advises that you indulge to the greatest extent possible, that your days on earth may be best spent. |
|||
From the beginning of complex formation, the pre-initiation complex is closed. |
|||
However, LaVey also cautions against failure to exercise self-control, and especially engaging in self-destructive behavior masked as "indulgence". This is commonly used as a Satanic argument against such things as drug use. LaVey also points out that religionist guilt preventing them from enjoying themselves is in fact only compulsion masked as religious piety, a compulsion to self-denial. |
|||
==Initiation== |
|||
'''IX.''' ''On the Choice of a Human Sacrifice'' |
|||
[[Image:simple transcription initiation1.svg|thumb|400px|Simple diagram of transcription initiation. RNAP = RNA polymerase II]] |
|||
In this essay, LaVey unequivocally condemns the practice of killing or harming an animal for ritual or magical purposes, in direct opposition to the common belief that Satanists advocate this practice. He states that to harm an innocent animal is a gross injustice and magically useless. |
|||
A collection of [[Transcription factor|transcription factors]] mediate the binding of RNA polymerase II and the initiation of transcription. Only after certain transcription factors are attached to the promoter does the polymerase bind to it. The completed assembly of transcription factors and RNA polymerase II (transcription initiation complex) bind to the promoter. |
|||
LaVey offers instead that the magician offer himself: in the case of a lust ritual, for example, through the production of an [[orgasm]]. He explains that the alleged purpose of an animal sacrifice is to release the vital energy of the animal to aid in the production of magic and that a superior magical effect is achieved by releasing one's own vital energy through orgasmic output. |
|||
Once the complex is opened by TFIIH initiation starts and the first phosphodiester bond is formed. |
|||
He also offers that while a Satanist does not under any circumstance advocate criminal activity, if you believe a person deserves to die, then you are perfectly justified in placing a curse upon them and making a figurative "human sacrifice" of them. |
|||
==Promoter Clearance== |
|||
While this essay does expound in important ways upon Satanic ethics, especially as regards the treatment of animals and certain aspects of magic, it is also wrought with a certain humor, as in the statement in reference to so-called "white magicians": |
|||
During the formation of the first nucleotide bond the RNA polymerase begins to clear the promoter. There is a tendency to release the RNA transcript and produce truncated transcripts (abortive initiation). Once the transcript reaches approximately 23 nucleotides it no longer slips and Elongation can occur. This is an ATP dependent process. The short-lived, unprocessed or partially processed, product is termed ''[[pre-mRNA]]''. |
|||
''One good orgasm would probably kill them!''<ref>LaVey, Anton ''The Satanic Bible'', pg 88</ref> |
|||
Promoter clearance also coincides with Phosphorylation of serine 5 on the carboxy terminal domain which is phosphorylated by TFIIH. |
|||
'''X.''' ''Life After Death Through Fulfillment of the Ego'' |
|||
== Eukaryotic pre-mRNA processing == |
|||
Taking further the non-spiritual view of the world, LaVey infers that if there is no afterlife, then this life must be valued very highly and the life of the individual should not be devalued. Therefore, he recommends that Satanists take great care to preserve their own lives so long as they can, and strongly criticizes the religious practice of martyrdom. |
|||
{{main|Post-transcriptional modification}} |
|||
Pre-mRNA requires extensive processing. |
|||
LaVey states that the circumstance under which a Satanist would willingly give up or risk his own life are very limited indeed. Among those circumstances he specifically names the defense of loved ones, especially one's spouse or children. LaVey views the defense of those you love as a natural instinct in animals (including man), and also as an informed risk: on the one hand, one's own life is of vital importance to oneself, but the life of those you love most may be equally important, and therefore a person may be forced in some circumstance to weigh this and may choose to defend those he loves with his own life. LaVey also condemns suicide, except under those circumstances where, as he puts it, life itself has become a form of abstinence and death has become an indulgence. This does not include those who suffer unwarranted self-loathing, but rather this refers to those who, due to terminal and painful disease or other such circumstance, cannot expect any more joy from their short life, ever, and chose to end it quickly and painlessly rather than endure ongoing suffering. |
|||
==== 5' cap addition ==== |
|||
Further, he takes the stance that as one will never achieve glory in the afterlife (as the afterlife does not exist), that one must strive for glory in life. He takes the somewhat classical stance that immortality is achieved by creating an enduring name for oneself by great deeds; compare this, for example, to myths of Greek heroes such as Achilles. |
|||
{{main|5' cap}} |
|||
A ''5' cap'' (also termed an RNA cap, an RNA 7-methylguanosine cap or an RNA m<sup>7</sup>G cap) is a modified guanine nucleotide that is added to the "front" or [[5' end]] of a the pre-mRNA using a 5',5-Triphosphate linkage shortly after the start of transcription. The 5' cap consists of a terminal 7-methylguanosine residue which is linked through a 5'-5'-triphosphate bond to the first transcribed nucleotide. Its presence is critical for recognition and proper attachment of mRNA by the [[ribosome]] and protection from 5' exonucleases [[RNase]]s. |
|||
'''XI.''' ''Religious Holidays'' |
|||
Cap addition is coupled to transcription, and occurs co-transcriptionally, such that each influences the other. Shortly after the start of transcription, the 5' end of the mRNA being synthesized is bound by a cap-synthesizing complex associated with [[RNA polymerase]]. This [[enzyme|enzymatic]] complex [[catalyze]]s the chemical reactions that are required for mRNA capping. Synthesis proceeds as a multi-step [[biochemistry|biochemical]] reaction. |
|||
LaVey briefly outlines the few Satanic holidays. |
|||
=== Coding regions === |
|||
The most important holiday in Satanism is one's own birthday, as the birth date of one's own god. To a Satanist, you are the most important being in the universe, and celebrating your own birthday honors your own vital existence. LaVey recommends that a Satanist celebrate his own birthday in any way he sees fit. The Satanic celebration of the birthday can also be seen as a mockery of the holidays commemorating the birth of various gods or saints in other religions. |
|||
{{main|Coding region}} |
|||
Coding regions are composed of [[codons]], which are decoded and translated into one protein by the ribosome. Coding regions begin with the [[start codon]] and end with the a [[stop codon| stop codons]]. Generally, the start codon is an AUG triplet and the stop codon is UAA, UAG, or UGA. The coding regions tend to be stabilised by internal base pairs, this impedes degradation.<ref>{{cite journal |author=Shabalina SA, Ogurtsov AY, Spiridonov NA |title=A periodic pattern of mRNA secondary structure created by the genetic code |journal=Nucleic Acids Res. |volume=34 |issue=8 |pages=2428–37 |year=2006 |pmid=16682450 |pmc=1458515 |doi=10.1093/nar/gkl287}}</ref><ref>{{cite journal |author=Katz L, Burge CB |title=Widespread selection for local RNA secondary structure in coding regions of bacterial genes |journal=Genome Res. |volume=13 |issue=9 |pages=2042–51 |year=2003 |month=September |pmid=12952875 |pmc=403678 |doi=10.1101/gr.1257503}}</ref> In addition to being protein-coding, portions of coding regions may serve as regulatory sequences in the [[pre-mRNA]] as [[exonic splicing enhancer]]s or [[exonic splicing silencer]]s. |
|||
After one's own birthday, LaVey names two other holidays of importance, [[Walpurgisnacht]] and [[Halloween]]. |
|||
=== Untranslated regions === |
|||
Chief among these holidays is [[Walpurgisnacht]], which in addition to the occult significance the date carries, also marks the formation of the Church of Satan in the year [[1966]], or ''I Annos Satanas''. This date is commonly celebrated by Satanists with private or group rituals, and private parties or family celebrations to commemorate the foundation of the Church of Satan. |
|||
{{main|5' UTR|3' UTR}} |
|||
Untranslated regions (UTRs) are sections of the mRNA before the start codon and after the stop codon that are not translated, termed the [[five prime untranslated region]] (5' UTR) and [[three prime untranslated region]] (3' UTR), respectively. These regions are transcribed with the coding region and thus are [[exon]]ic as they are present in the mature mRNA. Several roles in gene expression have been attributed to the untranslated regions, including mRNA stability, mRNA localization, and [[translational efficiency]]. The ability of a UTR to perform these functions depends on the sequence of the UTR and can differ between mRNAs. |
|||
[[Halloween]] is likewise celebrated for its occult significance, though Satanists tend to take a certain humor in its celebration, as it is also celebrated by non-Satanists. |
|||
The stability of mRNAs may be controlled by the 5' UTR and/or 3' UTR due to varying affinity for RNA degrading enzymes called [[ribonuclease]]s and for ancillary proteins that can promote or inhibit RNA degradation. |
|||
LaVey also names the summer and winter [[solstice]]s, and the spring and fall [[equinox]]es as Satanic holidays, as natural days of change in the seasons and days of universal ancient significance. |
|||
Translational efficiency, including sometimes the complete inhibition of translation, can be controlled by UTRs. Proteins that bind to either the 3' or 5' UTR may affect translation by influencing the ribosome's ability to bind to the mRNA. [[microRNA| MicroRNA]]s bound to the [[3' UTR]] also may affect translational efficiency or mRNA stability. |
|||
'''XII.''' ''The Black Mass'' |
|||
Cytoplasmic localization of mRNA is thought to be a function of the 3' UTR. Proteins that are needed in a particular region of the cell can actually be translated there; in such a case, the 3' UTR may contain sequences that allow the transcript to be localized to this region for translation. |
|||
The final essay of ''The Book of Lucifer'' begins the transition into the last two books of ''The Satanic Bible'' by a summary history of the "Black Mass", which LaVey outlines as being primarily a literary invention of Christians and inquisitors to impress upon people the depravity of the accused witches (which, as LaVey points out, were more often than not innocent of any witchcraft, guilty only of being eccentric, senile, or ugly). However, LaVey believes that emotionally evocative psychodrama has a place within Satanism as a means of emotional outlet and motivation, a topic he treats in detail in the remainder of ''The Satanic Bible''. |
|||
Some of the elements contained in untranslated regions form a characteristic [[secondary structure]] when transcribed into RNA. These structural mRNA elements are involved in regulating the mRNA. Some, such as the [[SECIS element]], are targets for proteins to bind. One class of mRNA element, the [[riboswitch]]es, directly bind small molecules, changing their fold to modify levels of transcription or translation. In these cases, the mRNA regulates itself. |
|||
The significance of ''The Book of Lucifer'' to Satanism cannot be overestimated. The topical essay format became LaVey's signature style, and the most important foundations of Satanic thought are contained within this section of ''The Satanic Bible''. Stylistically, it is carefully written with an economy of words to ensure that his points are always delivered clearly and not subject to reinterpretation, but at the same time restricted to only what needed to be said. It is also worth noting that LaVey refrains from any use of vulgarity in ''The Book of Lucifer'' and maintains a carefully worded, calm tone throughout in order that what he wrote here might be taken most seriously (although he does interject humor into his writing, even here). This contrasts with much later essays published in [[The Devil's Notebook]] and [[Satan Speaks!]], in which he writes more casually and freely. All of the essays contained in ''The Book of Lucifer'' are held to be invariable and indisputable Satanic dogma, whereas those later books contain a mixture of essays, some of which are considered dogma to Satanism, and others which are clearly personal opinions or musings by LaVey or simply essays on topics of interest to him. |
|||
==== Splicing ==== |
|||
''The Book of Lucifer'' is symbolically associated with the element of ''Air''. |
|||
{{main|Splicing (genetics)}} |
|||
Splicing is the process by which pre-mRNA is modified to remove certain stretches of non-coding sequences called [[intron]]s; the stretches that remain include protein-coding sequences and are called [[exon]]s. Sometimes pre-mRNA messages may be spliced in several different ways, allowing a single gene to encode multiple proteins. This process is called [[alternative splicing]]. Splicing is usually performed by an RNA-protein complex called the [[spliceosome]], but some RNA molecules are also capable of catalyzing their own splicing (''see [[ribozyme]]s''). |
|||
===The Book of Belial=== |
|||
''The Book of Belial: The Mastery of the Earth'' introduces in detail the Satanic concept of magic. It is, like the ''Book of Lucifer'', divided into essays, each of which brings greater explanation of what LaVey defined as magic and how he believed it could be applied. These essays and their meaning are briefly summarized here: |
|||
====Editing==== |
|||
'''I.''' ''The Theory and Practice of Satanic Magic (Definition and Purpose)'' |
|||
In some instances, an mRNA will be [[RNA editing|edited]], changing the nucleotide composition of that mRNA. In humans is the [[apolipoprotein B]] mRNA, which is edited in some tissues, but not others. The editing creates an early stop codon, which upon translation, produces a shorter protein. |
|||
LaVey gives the following definition for magic: |
|||
==== Polyadenylation ==== |
|||
''"The change in situations or events in accordance with one's will, which would, using normally accepted methods, be unchangeable".''<ref>LaVey, Anton ''The Satanic Bible'', pg 110</ref> |
|||
{{main|Polyadenylation}} |
|||
Polyadenylation is the covalent linkage of a polyadenylyl moiety to a messenger RNA molecule. Most messenger RNA (mRNA) molecules are polyadenylated at the 3' end. The [[messenger RNA#Poly(A) tail|poly(A) tail]] and the protein bound to it aid in protecting mRNA from degradation by exonucleases. Polyadenylation is also important for transcription termination, export of the mRNA from the nucleus, and translation. mRNA can also be polyadenylated in prokaryotic organisms, where poly(A) tails act to facilitate, rather than impede, exonucleolytic degradation. The 3' poly(A) tail is a long sequence of [[adenine]] nucleotides (often several hundred) at the [[3' end]] of the pre-mRNA. This tail promotes export from the nucleus and translation, and protects the mRNA from degradation. |
|||
LaVey then goes on to distinguish what he terms ''Lesser Magic'' from ''Greater Magic''. |
|||
=== Monocistronic versus polycistronic mRNA === |
|||
Lesser Magic consists of non-ritual or manipulative magic, through use of natural abilities to manipulate other humans and therefore circumstances by wile and guile. At the forefront of this effort, according to LaVey, is knowledge of how to employ appearances to one's advantage. He states that a person can employ contrived appearance to gain the alliance or obedience of others, and a competent magician can even combine these aesthetics as necessary. LaVey also states that a magician's actions to manipulate are an important component of Lesser Magic. LaVey later treated the matter of Lesser Magic in considerable detail in his book [[The Satanic Witch]]. |
|||
An mRNA molecule is said to be monocistronic when it contains the genetic information to [[Translation (genetics)|translate]] only a single [[protein]]. This is the case for most of the [[Eukaryote|eukaryotic]] mRNAs<ref name="Kozak_1983"> |
|||
{{ cite journal |
|||
| author = Kozak, M. |
|||
| year = 1983 |
|||
| month = March |
|||
| title = Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles |
|||
| journal = Microbiological Reviews |
|||
| volume = 47 |
|||
| issue = 1 |
|||
| pages = 1–45 |
|||
| doi = |
|||
| pmid = 6343825 |
|||
| url = http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=281560&blobtype=pdf |
|||
| format = PDF |
|||
| accessdate = 2006-08-12 |
|||
}} |
|||
</ref>. |
|||
==Elongation== |
|||
Greater Magic includes all ritual and ceremonial magic, which LaVey spends the remainder of ''The Satanic Bible'' detailing. |
|||
[[Image:simple transcription elongation1.svg|thumb|400px|Simple diagram of transcription elongation]] |
|||
'''II.''' ''The Three Types of Satanic Ritual'' |
|||
One strand of DNA, the ''template strand'' (or non-coding strand), is used as a template for RNA synthesis. As transcription proceeds, RNA polymerase II traverses the template strand and uses base pairing complementarity with the DNA template to create an RNA copy. Although polymerase traverses the template strand from 3' → 5', the coding (non-template) strand is usually used as the reference point, so transcription is said to go from 5' → 3'. This produces an RNA molecule from 5' → 3', an exact copy of the coding strand (except that [[thymine]]s are replaced with [[uracil]]s, and the nucleotides are composed of a ribose (5-carbon) sugar where DNA has deoxyribose (one less oxygen atom) in its sugar-phosphate backbone). |
|||
LaVey names three types of Satanic ritual: |
|||
mRNA transcription can involve multiple RNA polymerases on a single DNA template and multiple rounds of transcription (amplification of particular mRNA), so many mRNA molecules can be produced from a single copy of a gene. This step also involves a proofreading mechanism that can replace incorrectly incorporated bases. |
|||
Lust Rituals are conducted for the purpose of sexually attracting a person of your choice. LaVey specifies that you must have a particular person, or at least type of person, in mind for this to have any chance of success. |
|||
The polymerase can experience pauses. These pauses may be intrinsic to RNA polymerase II or due to chromatin structure. Often the polymerase pauses to allow appropriate RNA editing factors to bind. |
|||
Compassion Rituals are performed for the gain of those you care for, or on one's own behalf. The purpose is to increase worldly gain for the target, whether it be a friend or yourself. Any ritual aimed at gaining material wealth, physical advantage, or increase in life station falls into this category. |
|||
==Termination== |
|||
Destruction Rituals are otherwise known as curses or hexes, and are employed for the destruction of one's enemies. |
|||
[[Image:simple transcription termination1.svg|thumb|400px|Simple diagram of transcription termination]] |
|||
LaVey also warns that in each of these cases, the only risk is that you must truly want to see what you have wished for come to pass. He very clearly states that there is no guilt-ridden risk that your rituals (specifically, destruction rituals) will be returned upon you (such as the "[[Rule of Three (Wiccan)|threefold rule]]"), but rather that you must be aware of the consequences should you get what you want. He advises that if you perform a lust ritual, that you be prepared to take what you have desired should it come to you; if you perform a compassion ritual, be aware that all gains may be at another's expense; if you perform a destruction ritual, that you should not care whether your enemy lives or dies. |
|||
Transcription termination in eukaryotes is less well understood, but involves cleavage of the new transcript, followed by template-independent addition of ''A''s at its new 3' end, in a process called [[polyadenylation]]. |
|||
'''III.''' ''The Ritual, or "Intellectual Decompression Chamber"'' |
|||
Polyadenylation occurs during and immediately after transcription of DNA into RNA. After transcription has been terminated, the mRNA chain is cleaved through the action of an endonuclease complex associated with RNA polymerase II. After the mRNA has been cleaved, around 250 adenosine residues are added to the free 3' end at the cleavage site. This reaction is catalyzed by polyadenylate polymerase. Just as in alternative splicing, there can be more than one polyadenylation variant of a mRNA. |
|||
LaVey begins by explaining the role of both solitary and group rituals, and which kinds of rituals are suited to group performance and which are not. He suggests that destruction rituals can be enhanced by group participation, but that compassion and lust rituals, due to their highly personal nature, are best performed alone, as self-consciousness has no place in the ritual chamber. |
|||
Once completely processed, the mRNA is termed ''[[mature mRNA]]''. |
|||
He then describes the ritual chamber as an "Intellectual Decompression Chamber", or a means of releasing pent up energy by willfully entering into a state of conscious suspension of disbelief. He adds that only by relieving oneself of intellectual critique of what one is doing in the ritual chamber, can one hope to truly achieve magical ends. He acknowledges that this is similar in principle to the rituals of other religions, but claims a distinction from them, as Satanists are consciously aware of what they are doing and the fact that they are entering into a suspension of disbelief for specific purposes, instead of the self-deceit and delusion characteristic of other religions. |
|||
== Structure == |
|||
'''IV.''' ''The Ingredients Used in the Performance of Satanic Magic'' |
|||
[[Image:MRNA structure.png|thumb|700px|center|The structure of a mature eukaryotic mRNA. A fully processed mRNA includes a [[5' cap]], [[5' UTR]], [[coding region]], [[3' UTR]], and poly(A) tail.]] |
|||
=Transport= |
|||
LaVey names five elements essential to a magical working: |
|||
Because eukaryotic transcription and translation is compartmentally separated, eukaryotic mRNAs are exported from the [[cell nucleus|nucleus]] to the [[cytoplasm]]. Mature mRNAs are recognized by their processed modifications and then exported through the [[nuclear pore]]. |
|||
''Desire'': The magician must possess great desire to see the intended outcome come to fruition. |
|||
=Translation= |
|||
''Timing'': A time for ritualization should be chosen to align with whatever time the target is most receptive; LaVey especially names the period in which the target is in deep sleep as the ideal time for this. |
|||
{{main|Translation (genetics)}} |
|||
Eukaryotic mRNA that has been processed and transported to the cytoplasm (i.e. mature mRNA) can then be translated by the ribosome. Translation may occur at [[ribosomes]] free-floating in the cytoplasm, or directed to the [[endoplasmic reticulum]] by the [[signal recognition particle]]. |
|||
''Imagery'': Accoutrements conducive to the ritual environment, and the full visualization of the desired outcome, must be present. This not only includes the standard ritual equipment, but more specifically any specialized imagery or items the magician requires to give him a full mental view of what he wishes to happen. This can include drawings or paintings, sculptures, dolls, written poems or verses, or anything else that aids in visualizing the outcome. |
|||
=[[Post-translational modification]]= |
|||
''Direction'': As mentioned before, the magician must have a very clear target in mind. All three types of ritual demand that the magician know specifically who (or at least, what kind of person) he wishes to be targeted by his magic. The magician must also be able to give vent to all his desires during the ritual, not before or after. |
|||
This may include the formation of [[disulfide bridge]]s or attachment of any of a number of biochemical [[functional group]]s, such as [[acetate]], [[phosphate]], various [[lipid]]s and [[carbohydrate]]s. [[Enzyme]]s may also remove one or more amino acids from the leading (amino) end of the polypeptide chain, leaving a protein consisting of two polypeptide chains connected by disulfide bonds. |
|||
''Balance'': The magician must temper his magic with a dose of common sense, otherwise known as the balance factor. LaVey states that ritualized desires must be realistic; wishing for the impossible or the absurdly far-fetched will not yield results, as the magician cannot reasonably hope to put forth enough magical energy to accomplish what cannot be accomplished by any means. To quote LaVey: |
|||
=[[Protein folding]]= |
|||
''Magic is like nature itself, and success in magic requires working in harmony with nature, not against it.''{{Fact|date=November 2007}} |
|||
During and after synthesis, polypeptide chains often fold to assume, so called, native [[secondary structure|secondary]] and [[tertiary structure]]s. |
|||
'''V.''' ''The Satanic Ritual'' |
|||
=Degradation= |
|||
This segment of ''The Book of Belial'' begins detailed instructions for actual performance of ritual and how it is conducted. It includes instructions of selection and use of ritual attire, the altar, [[Sigil of Baphomet]], candles, bell, chalice, elixir, sword, phallus, gong, and parchment. |
|||
After a certain amount of time, the message is degraded by [[RNase]]s. The limited lifetime of mRNA enables a cell to alter protein synthesis rapidly in response to its changing needs. |
|||
The ''Book of Belial'' is symbolically associated with the element of [[earth]]. |
|||
Different mRNAs within the same cell have distinct lifetimes (stabilities). In mammalian cells, mRNA lifetimes range from several minutes to days. The greater the stability of an mRNA, the more protein may be produced from that mRNA. The presence of [[AU-rich element]]s in some mammalian mRNAs tends to destabilize those transcripts through the action of cellular proteins that bind these motifs. Rapid mRNA degradation via [[AU-rich element]]s is a critical mechanism for preventing the overproduction of potent cytokines such as tumor necrosis factor (TNF) and granulocyte-macrophage colony stimulating factor (GM-CSF).<ref name="Shaw1986">{{cite journal |author=Shaw G, Kamen R |title=A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation |journal=Cell |volume=46 |issue=5 |pages=659–67 |year=1986 |month=August |pmid=3488815 |doi=10.1016/0092-8674(86)90341-7}}</ref> Base pairing with a small interfering RNA ([[siRNA]]) or microRNA ([[miRNA]]) can also accelerate mRNA degradation. |
|||
===The Book of [[Leviathan]]=== |
|||
''The Book of Leviathan: The Raging Sea'' contains explicit instructions as to what is to be said and done during ritual. Its text is largely comprised of invocations and ritual verse, including the ''Invocation to Satan'' performed at the outset of each ritual, ''[[The infernal names|The Infernal Names]]'', and separate invocations for each of the three ritual types. It also includes the nineteen [[Enochian]] Keys, a dark reinterpretation of [[John Dee (mathematician)|John Dee]]'s verses of the same name in the fictive language Enochian. These keys serve as moving ritual chants in Satanic ritual, and the English translations serve likewise as versed statements of Satanic dogma. The meaning of the Enochian Keys was altered by LaVey from John Dee's version in an effort to make them more consistent with Satanic dogma while retaining their usefulness as ritual devices. |
|||
=References= |
|||
The Book of Leviathan is symbolically associated with the element of water (as opposed to fire). |
|||
{{Reflist|2}} |
|||
=Further reading= |
|||
==Yankee Rose== |
|||
{{Cleanup-section|date=March 2007}} |
|||
''The Satanic Bible'' ends with a mysterious stamp reading only "YANKEE ROSE", in all capital letters, and in slightly inkier typing. The meaning of this is a matter of speculation, and remains a mystery, to both the public and Satanists alike. The [[Church of Satan]] website, however, offers one possible clue in the song by that name, recorded by LaVey on his record [[Satan Takes a Holiday]]. However, the website also adds that this may or may not be a clue at all. LaVey wished its meaning to remain secret, and the true significance of "Yankee Rose" shall forever remain a mystery.<ref>[http://www.churchofsatan.com/Pages/YankeeRose.html The Enigma of Yankee Rose<!-- Bot generated title -->]</ref> |
|||
* Doolittle, R.F. (1989) Redundancies in protein sequences. In ''Predictions of Protein Structure and the Principles of Protein Conformation'' (Fasman, G.D. ed) Plenum Press, New York, pp. 599-623 |
|||
There have been many rumors over the years regarding the meaning of the words "YANKEE ROSE" in this context, including claims that they form an [[anagram]] or is a form of [[cryptogram]], the name of a haunted ship, or even the brand name of the organ LaVey played in the circus (a period of his life where he made so many of the observations that eventually went into ''The Satanic Bible''). |
|||
* David L. Nelson and Michael M. Cox, ''Lehninger Principles of Biochemistry'', 3rd edition, 2000, Worth Publishers, ISBN 1-57259-153-6 |
|||
* [[Uwe Meierhenrich]], ''Amino acids and the asymmetry of life'', Springer-Verlag, 2008, ISBN 978-3-540-76885-2 |
|||
=See also= |
|||
However, LaVey did record the Holden/Frankl song of the same title as part of a medley on his "[[Satan Takes a Holiday]]" album, initially released by [[Amarillo Records]] and since re-released by [[Reptilian Records]] on their Adversary label. Some say this jaunty tune was one LaVey used to end his sets in his early days of playing organ for bars and nightclubs. |
|||
*[[Genetic code]] |
|||
*[[Cistron]] |
|||
*[[Operon]] |
|||
*[[lac operon|''lac'' operon]] |
|||
=External links= |
|||
LaVey once wrote about using old pop music in Satanic rituals: "I have my personal favorites which are readily identifiable with meaningful situations. Perhaps one day I will share them with you, and it will be seen that many [[Satanists]] favor the same tunes!"<ref>[http://archives.stupidquestion.net/sq21700.html Stupid Question <!-- Bot generated title -->]</ref> He also talks about such music in several other writings and interviews. |
|||
==Amino Acids== |
|||
It's therefore widely believed that the phrase is a reference to the song, though it's still not entirely known why LaVey decided to include the phrase in the book. |
|||
* [http://isoelectric.ovh.org Protein isoelectric point according amino acids charge] |
|||
David Lee Roth's song "[[Yankee Rose (song)|Yankee Rose]]" came out around the time of the "[[Satanic Panic]]" hysteria of the 1980s, a time when many rock bands were falsely accused of practicing or promoting devil worship, or including [[subliminal message]]s on their albums. And the fact that the phrase "Yankee Rose" shows up in ''The Satanic Bible'' didn't help Roth in this regard. However, again, the use of the same words here is purely coincidental. |
|||
* [http://www.mathiasbader.de/studium/biology/index.php?lng=en Learn the 20 proteinogenic amino acids online] |
|||
* [http://www.unc.edu/~bzafer/aminoacids/ListOfStandardAminoAcids.pdf The PDF List of Standard Amino Acids] |
|||
* [http://micro.magnet.fsu.edu/aminoacids/index.html Molecular Expressions: The Amino Acid Collection] - Has detailed information and microscopy photographs of each amino acid. |
|||
* [http://www.chem.qmul.ac.uk/iupac/AminoAcid/ Nomenclature and Symbolism for Amino Acids and Peptides] International Union of Pure and Applied Chemistry and The International Union of Biochemistry and Molecular Biology. IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN) |
|||
* [http://www.peptideguide.com/amino-acids/index.html Amino acids overview at Peptide Guide] |
|||
* [http://www.russell.embl.de/aas/ Amino acid properties] - Properties of the amino acids (a tool aimed mostly at molecular geneticists trying to understand the meaning of mutations) |
|||
* [http://www.newscientist.com/article.ns?id=mg19025545.200&feedId=online-news_rss20 Right-handed amino acids were left behind] |
|||
* [http://www2.iq.usp.br/docente/gutz/Curtipot_.html Amino acid solutions pH, titration curves and distribution diagrams - freeware] |
|||
* [http://www.organic-chemistry.org/synthesis/C1C/nitrogen/alpha-amino-acids2.shtm Synthesis of Amino Acids and Derivatives] |
|||
* [http://researchnews.osu.edu/archive/aminoacd.htm 22nd amino acid] - Press release from Ohio State describing the discovery of pyrrolysine as a 22nd amino acid. |
|||
*[http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=stryer.TOC&depth=2 NCBI Bookshelf Free Textbook Access] |
|||
==Nucleotide synthesis== |
|||
==The Satanic Rituals== |
|||
{{main|The Satanic Rituals}} |
|||
* [http://www.chem.qmul.ac.uk/iubmb/enzyme/reaction/misc/pyrimid.html Overview] at [[Queen Mary, University of London]] |
|||
'''''[[The Satanic Rituals]]''''' was published as a companion edition to '''''[[The Satanic Bible]]''''', and contains a number of other rituals, many of them very elaborate. Both books are published by Avon and remain in print. |
|||
==Protein synthesis== |
|||
* [http://www.accessexcellence.org/AB/GG/protein_synthesis.html Protein Synthesis] |
|||
* [http://learningobjects.wesleyan.edu/proteinsynthesis/ Protein Synthesis Animation] Wesleyan University Learning Objects animation of protein synthesis. |
|||
* [http://www.scienceaid.co.uk/biology/genetics2/proteinsynthesis.html Science aid:Protein synthesis] For high school |
|||
==Transcription== |
|||
* [http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/T/Transcription.html Transcription] |
|||
==References== |
|||
* [http://cmol.nbi.dk/models/dynamtrans/dynamtrans.html Interactive Java simulation of transcription initiation.] From [http://cmol.nbi.dk/ Center for Models of Life] at the Niels Bohr Institute. |
|||
<references/> |
|||
* From RNA to Protein Synthesis [http://www.asterpix.com/console/?avi=7698391 hypervideo]. |
|||
== |
==Translation== |
||
*[http://www.churchofsatan.com Church of Satan Website |
|||
*: [http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/T/Translation.html Translation] |
|||
{{DEFAULTSORT:Satanic Bible, The}} |
|||
* [http://www.ncbi.nlm.nih.gov/books/bv.fcgi?highlight=translation&rid=cooper.section.1167#1178 Translation of mRNA] Section of ''The Cell: A Molecular Approach'' by Geoffrey M. Cooper |
|||
[[Category:1969 books]] |
|||
[[Category:Satanic texts]] |
|||
[[Category:Philosophy books]] |
|||
{{AminoAcids}} |
|||
{{Amino acid metabolism enzymes}} |
|||
{{MetabolismMap}} |
|||
{{Metabolism-stub}} |
|||
{{Nucleotide metabolism}} |
|||
{{Protein biosynthesis}} |
|||
{{Protein metabolism}} |
|||
{{Protein primary structure}} |
|||
{{Protein topics}} |
|||
[[Category:Chemical synthesis]] |
|||
[[af:The Satanic Bible]] |
|||
[[Category:Gene expression]] |
|||
[[cs:Satanská bible]] |
|||
[[Category:Metabolism]] |
|||
[[da:Satans bibel]] |
|||
[[Category:Proteins]] |
|||
[[de:Satanische Bibel]] |
|||
[[Category:Protein biosynthesis]] |
|||
[[et:Saatanlik Piibel]] |
|||
[[Category:Pyrimidines]] |
|||
[[eo:Satana Biblio]] |
|||
[[fa:انجیل شیطانی]] |
|||
[[fr:Bible satanique]] |
|||
[[hr:Sotonistička Biblija]] |
|||
[[he:התנ"ך השטני]] |
|||
[[lv:Sātaniskā bībele]] |
|||
[[lt:Šėtono biblija]] |
|||
[[nl:Satansbijbel]] |
|||
[[pl:Biblia Szatana]] |
|||
[[ru:Сатанинская библия]] |
|||
[[sk:Satanská biblia]] |
|||
[[fi:Saatanallinen Raamattu]] |
|||
[[sv:Den Satanistiska Bibeln]] |
|||
[[tr:Şeytani İncil]] |
|||
Revision as of 02:00, 14 October 2008
Biosynthesis of a Human Protein
| Human Lineage | |
|---|---|
| Scientific classification | |
| Domain: | |
| Kingdom: | |
| Phylum: | |
| Subdivision: | |
| Subphylum: | |
| Infraphylum: | |
| Class: | |
| Infraclass: | |
| Superorder: | |
| Order: | |
| Suborder: | |
| Infraorder: | |
| Parvorder: | É. Geoffroy, 1812
|
| Family: | |
| Subfamily: | |
| Tribe: | |
| Subtribe: | |
| Genus: | Linnaeus, 1758
|
| Species: | sapiens
|
| Subspecies: | sapiens
Homo sapiens sapiens
|
Consider starting with a specific gene. Assemble the transcription mechanism at the appropriate genome location. Transcribe the gene using nucleotide accountability.
Protein biosynthesis
A protein is a naturally occurring aliphatic compound, composed of amino acids (aa) and containing primarily at least the elements carbon (C), hydrogen (H), nitrogen (N), and oxygen (O). Many proteins also contain sulfur (S), occasionally phosphorus (P), and some iron (Fe).
Amino acids combine in a condensation reaction that releases water (dehydration reaction) to form a residue held together by a peptide bond. The digestion of dietary proteins produces dipeptides which are absorbed more rapidly than aa. Some tripeptides and tetrapeptides are synthesized in humans. Proteins are polypeptide molecules. Each protein is defined by a unique sequence of amino acid residues. This sequence is the primary structure of the protein.
In biosynthesis, precursors are activated often with nucleotides, generally in the presence of a catalyst, oxidizing or reducing agent, to interconvert biochemicals within living organisms.
Amino acid synthesis
Metabolic pathways) produce amino acids from other compounds. The substrates for these pathways are usually ingested or synthesized internally by intestinal microbiota or activation of the heterochromatin. Of the proteinogenic aa, humans are able to synthesise up to 12 at the level needed for normal growth. The remaining essential aa must be supplied with or from food.[1]
| Essential | Nonessential |
|---|---|
| Isoleucine | Alanine |
| Leucine | Asparagine |
| Lysine | Aspartate |
| Methionine | Cysteine* |
| Phenylalanine | Glutamate |
| Threonine | Glutamine* |
| Tryptophan | Glycine* |
| Valine | Proline* |
| Serine* | |
| Tyrosine* | |
| Arginine* | |
| Histidine* |
(*) Essential only in certain cases.[2][3]
The situation is a more complicated since cysteine, tyrosine, histidine and arginine are semiessential aa in children. Their metabolic pathways are not fully developed.[4] The aa amounts required depend on the age and health of the individual.
In the citric acid cycle glutamate dehydrogenase catalyzes the reductive amination of α-ketoglutarate to glutamate:
At this step, the chirality of the amino acid is established. Afterwards, alanine and aspartate are synthesized by the transamination of glutamate, vis a vis the transamination of pyruvate and oxaloacetate, respectively. All of the remaining amino acids are constructed from glutamate or aspartate, by transamination of these two amino acids with one α-keto acid. Glutamine is synthesized from NH4+ and glutamate, and asparagine is synthesized similarly. Proline and arginine are derived from glutamate. Serine formed from 3-phosphoglycerate, is the precursor of glycine and cysteine. Tyrosine is synthesized by the hydroxylation of phenylalanine, an essential amino acid.
Most of the pathways of amino acid biosynthesis are regulated by feedback inhibition, in which the committed step is allosterically inhibited by the final product. Branched pathways require extensive interaction among the branches that includes both negative and positive regulation. The regulation of glutamine synthetase from E. coli is a striking demonstration of cumulative feedback inhibition and of control by a cascade of reversible covalent modifications.
Nucleotide synthesis
De novo synthesis
Purine nucleotides

Pyrimidine nucleotides
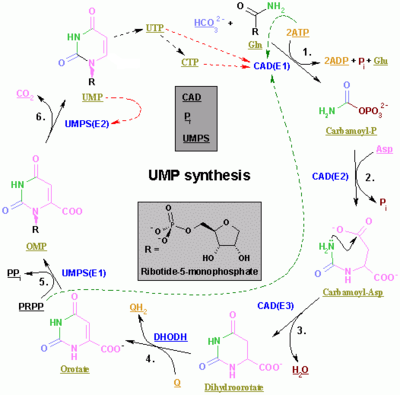
Salvage synthesis
Adenosine triphosphate (ATP)
Anaerobic respiration
Glucose C6H12O6 2 C3H6O3 Lactic acid + 2 ATP.
De novo synthesis of purine nucleotides
Salvage synthesis
- Adenine + Phosphoribosyl pyrophosphate PRPP <----> AMP + PPi : Enzyme (Adenine phosphoribosyltransferase) APRT[5], GeneID: 353.
Cytidine triphosphate (CTP)
Uridine 5'-triphosphate (UTP) + Glutamine + ATP + H2O => Cytidine 5'-triphosphate + Glutamate + ADP + Orthophosphate (Pi) : Enzyme (Cytidine 5' triphosphate synthetase) CTPS, GeneID: 1503.
Guanosine triphosphate (GTP)
Uridine triphosphate (UTP)
De novo synthesis of the pyrimidine ring and conversion to UMP:
- L-Glutamine (Gln) + 2 ATP + HCO3- + H2O => Carbamoyl phosphate + 2 ADP + Orthophosphate (Pi) + Glutamic acid (Glu) : Enzyme 1 (Carbomoyl-phosphate Synthetase 2, CPS II) CAD(E1), GeneID: 790.
- Carbamoyl phosphate + L-Aspartate <=> N-carbamoyl L-aspartate + Orthophosphate (Pi) : Enzyme 2 (Aspartate transcarbamylase, ATCase) CAD(E2), GeneID: 790.
- N-Carbamoyl L-Aspartate + H+ <=> (S)-Dihydroorotate + H2O : Enzyme 3 (Dihydroorotase, DHO) CAD(E3), GeneID: 790.
- (S)-Dihydroorotate + Ubiquinone => Orotate + Ubiquinol : Enzyme (Dihydroorotate dehydrogenase) DHODH, GeneID: 1723
- Orotate + 5-Phospho-alpha-D-Ribose 1-Diphosphate (PRPP) <=> Orotidine 5'-monophosphate (OMP) + PPi : Enzyme 1 (Uridine monophosphate synthetase) UMPS(E1), GeneID: 7372.
- Orotidine 5'-monophosphate => Uridine 5'-monophosphate (UMP) + CO2 : Enzyme 2 UMPS(E2), GeneID: 7372
- Uridine 5'-monophosphate (UMP) + ATP <=> Uridine 5'-diphosphate (UDP) + ADP : Enzymes (Uridine monophosphate kinase) UMPK, GeneID: 51727; (uridine-cytidine kinase 2) UCK2, GeneID:7371.
- Uridine 5'-diphosphate (UDP) + ADP <=> uridine 5'-triphosphate (UTP) + AMP : Enzymes (Nucleoside-diphosphate kinases A, B, C, D; NME6, NME7) NDKA-D, GeneIDs: 4830, 4831, 4832, 4833; NME6 GeneID: 10201, NME7 GeneID: 29922.
Transcription
A DNA sequence is enzymatically copied by RNA polymerase II to produce a complementary nucleotide RNA strand of messenger RNA (mRNA). mRNA is modified through RNA splicing, 5' end capping, and the addition of a polyA tail.[6]
Transcriber
In human biochemistry a transcriber might be thought of as a tool assembled in the nucleus for the transcription of genetic information. It synthesizes (a nucleic acid, usually RNA) using an existing nucleic acid (DNA) as a template from which to copy.
Transcription factors
Transcription factors bind to specific sequences of DNA and are part of the system that controls the transcription of genetic information from DNA to RNA.[7][8] Transcription factors occur in a complex, increase (as an activator), or prevent (as a repressor) the presence of RNA polymerase II.[9][10][11]
Each transcription factor contains one or more DNA binding domains (DBDs) which attach to specific sequences of DNA adjacent to the gene that it regulates.[12][13]
There are approximately 2600 proteins in the human genome that contain DNA-binding domains and most of these are presumed to function as transcription factors.[14]
The combinatorial use of a subset of the approximately 2000 human transcription factors easily accounts for the unique regulation of each gene in the human genome during development.[15]
General transcription factors (GTFs) are necessary for transcription to occur. GTFs are part of the large transcription preinitiation complex that interacts with RNA polymerase II directly.[16] They are ubiquitous and interact with the core promoter region surrounding the transcription start site(s) of allclass II genes.[17]
TFIIA TFIIB TFIID (see also TATA binding protein) TFIIE TFIIF TFIIH
Active transcription units (transcription factors) are clustered in the nucleus. There are ~10,000 factors in the nucleoplasm of a HeLa cell, among which are ~8,000 polymerase II factors and ~2,000 polymerase III factors. Each polymerase II factor contains ~8 polymerases. As most active transcription units are associated with only one polymerase, each factor will be associated with ~8 different transcription units. These units might be associated through promoters and/or enhancers, with loops forming a ‘cloud’ around the factor.
Additional proteins: coactivators chromatin remodelers histone acetylases deacetylases kinases methylases
play crucial roles in gene regulation.[15]
The TATA binding protein (TBP, GeneID: 6908) is a transcription factor that binds specifically to a DNA sequence called the TATA box. This DNA sequence is found about 25-30 base pairs upstream of the transcription start site in some eukaryotic gene promoters.[18] TBP, along with a variety of TBP-associated factors, make up the TFIID, a general transcription factor that in turn makes up part of the RNA polymerase II preinitiation complex.[11] As one of the few proteins in the preinitation complex that binds DNA in a sequence-specific manner, it helps position RNA polymerase II over the transcription start site of the gene. However, it is estimated that only 10-20% of human promoters have TATA boxes. Therefore, TBP is probably not the only protein involved in positioning RNA polymerase II.
TBP is a subunit of the eukaryotic transcription factor TFIID. TFIID is the first protein to bind to DNA during the formation of the pre-initiation transcription complex of RNA polymerase II (RNA Pol II). Binding of TFIID to the TATA box in the promoter region of the gene initiates the recruitment of other factors required for RNA Pol II to begin transcription. Some of the other recruited transcription factors include TFIIA, TFIIB and TFIIF. Each of these transcription factors are formed from the interaction of many protein subunits, indicating that transcription is a heavily regulated process.
When TBP binds to a TATA box within the DNA, it distorts the DNA by inserting amino acid side chains between base pairs, partially unwinding the helix, and doubly kinking it. The distortion is accomplished through a great amount of surface contact between the protein and DNA. TBP binds with the negatively charged phosphates in the DNA backbone through positively charged lysine and arginine amino acid residues. The sharp bend in the DNA is produced through projection of four bulky phenylalanine residues into the minor groove. As the DNA bends, its contact with TBP increases, thus enhancing the DNA-protein interaction.
The strain imposed on the DNA through this interaction initiates melting, or separation, of the strands. Because this region of DNA is rich in adenine and thymine residues, which base pair through only two hydrogen bonds, the DNA strands are more easily separated. Separation of the two strands exposes the bases and allows RNA polymerase II to begin transcription of the gene.
RNA polymerase II
Each of the RNA polymerases transcribes different classes of proteins. Specifically, RNA polymerase I transcribes rRNA and RNA polymerase III transcribes various additional RNAs.
RNA polymerase II transcribes Class II genes to form proteins. It catalyzes the transcription of DNA to synthesize precursors of mRNA and most snRNA and microRNA.[19]
It is a 550 kDa complex of 12 subunits:
Polymerase (RNA) II (DNA directed) polypeptide A, 220kDa (POLR2A), GeneID: 5430.
This largest subunit contains a carboxy terminal domain composed of heptapeptide repeats that are essential for polymerase activity. These repeats contain serine and threonine residues that are phosphorylated in actively transcribing RNA polymerase. In addition, this subunit, in combination with several other polymerase subunits, forms the DNA binding domain of the polymerase, a groove in which the DNA template is transcribed into RNA.[20]
Polymerase (RNA) II (DNA directed) polypeptide B, 140kDa (POLR2B), GeneID: 5431.
This second largest subunit, in combination with at least two other polymerase subunits, forms a structure within the polymerase that maintains contact in the active site of the enzyme between the DNA template and the newly synthesized RNA.[21]
Polymerase (RNA) II (DNA directed) polypeptide C, 33kDa (POLR2C), GeneID: 5432.
This third largest subunit contains a cysteine rich region and exists as a heterodimer with another polymerase subunit, POLR2J. These two subunits form a core subassembly unit of the polymerase.[22]
DNA-directed RNA polymerase II polypeptide D (or subunit B4) (POLR2D), GeneID: 5433.
This is the fourth largest subunit.[23]
Polymerase (RNA) II (DNA directed) polypeptide E, 25kDa (POLR2E), GeneID: 5434.
This the fifth largest subunit is present in two-fold molar excess over the other polymerase subunits. An interaction may occur between transcriptional activators and the polymerase through this subunit.[24]
Polymerase (RNA) II (DNA directed) polypeptide F (POLR2F), GeneID: 5435.
This sixth largest subunit, based on studies in yeast, in combination with at least two other subunits, may form a structure that stabilizes the transcribing polymerase on the DNA template.[25]
Polymerase (RNA) II (DNA directed) polypeptide G (POLR2G), GeneID: 5436.
This seventh largest subunit, from studies in yeast, may play a role in regulating polymerase function.[26]
Polymerase (RNA) II (DNA directed) polypeptide H (POLR2H), GeneID: 5437.
This is an essential subunit.[27]
Polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa (POLR2I), GeneID: 5438.
This subunit in combination with two other polymerase subunits forms the DNA binding domain of the polymerase, a groove in which the DNA template is transcribed into RNA. The product of this gene has two zinc finger motifs with conserved cysteines and the subunit does possess zinc binding activity.[28]
Polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa (POLR2J), GeneID: 5439.
This subunit exists as a heterodimer with another polymerase subunit; forming a core subassembly unit of the polymerase. Two similar genes are located nearby on chromosome 7q22.1.[29]
DNA directed RNA polymerase II polypeptide J-related gene (POLR2J2), GeneID: 246721.[30]
This gene is a member of the RNA polymerase II subunit 11 gene family, which includes three genes in a cluster on chromosome 7q22.1. The founding member of this family, DNA directed RNA polymerase II polypeptide J, encodes multiple, alternatively spliced transcripts that potentially express isoforms with distinct C-termini compared to DNA directed RNA polymerase II polypeptide J. Most or all variants are spliced to include additional non-coding exons at the 3' end which makes them candidates for nonsense-mediated decay (NMD). Consequently, it is not known if this locus expresses a protein or proteins in vivo.[31]
DNA directed RNA polymerase II polypeptide J3 (POLR2J3), GeneID: 548644.[32]
This gene is a member of the RNA polymerase II subunit 11 gene family, which includes three genes in a cluster on chromosome 7q22.1. This locus produces multiple, alternatively spliced transcripts that potentially express isoforms with distinct C-termini compared to DNA directed RNA polymerase II polypeptide J. Most or all variants are spliced to include additional non-coding exons at the 3' end which makes them candidates for nonsense-mediated decay (NMD). Consequently, it is not known if this locus expresses a protein or proteins in vivo.[33]
DNA directed RNA polymerase II polypeptide J4, pseudogene (POLR2J4), GeneID: 84820.[34]
Although now thought to be a pseudogene, model predictions suggests that a miscRNA from this gene of possible transcription function may be involved in DNA binding and DNA-directed RNA polymerase activity.[35]
Polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa (POLR2K), GeneID: 5540.
This is one of the smallest subunits.[36]
RP11-90M2.3 novel protein similar to Polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa (RP11-90M2.3), GeneID: 100129174.
This maybe another small subunit.[37]
Polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa (POLR2L), GeneID: 5441.
The subunit contains four conserved cysteines characteristic of an atypical zinc-binding domain.[38]
Initiation Regulation
Initiation is regulated by several mechanisms:
- Protein interference.
- Chromatin structure inhibition.
- Phosphorylation.
Regulation by Protein interference
Protein interference is the process where some signaling protein interacts, either with the promoter or some stage of the partially constructed complex, to prevent further construction of the polymerase complex. This is generally a very rapid response and is used for fine level, individual gene control and for 'cascade' processes for a group of genes useful under a specific conditions (for example DNA repair genes or heat shock genes).
Regulation by Chromatin structure inhibition
Chromatin structure inhibition is the process where the promoter is hidden by chromatin structure. Chromatin structure is controlled by post-translational modification of the histones involved and leads to gross levels of high or low transcription levels. See: chromatin, histone and nucleosome.
These methods of control can be combined in a modular method, allowing very high specificity in transcription initiation control.
Regulation by Phosphorylation
POLR2A has a domain at its C-terminus, the CTD (C-terminal domain) which is the target of kinases and phosphatases. The phosphorylation of the CTD is an important regulation mechanism, as this allows attraction and rejection of factors that have a function in the transcription process. The CTD can be considered as a platform for transcription factors.
The CTD consists of repetitions of an amino acid motif, YSPTSPS, of which Serines and Threonines can be phosphorylated. The mammalian protein contains 52. There are many different combinations of phosphorylations possible on these repeats and these can change rapidly during transcription. The regulation of these phosphorylations and the consequences for the association of transcription factors plays a major role in the regulation of transcription.
During the transcription cycle, the CTD of POLR2A is reversibly phosphorylated. RNAP II containing unphosphorylated CTD is recruited to the promoter, whereas the hyperphosphorylated CTD form is involved in active transcription. Phosphorylation occurs at two sites within the heptapeptide repeat, at Serine 5 and Serine 2. Serine 5 phosphorylation is confined to promoter regions and is necessary for the initiation of transcription, whereas Serine 2 phosphorylation is important for mRNA elongation and 3'-end processing.
Mediator (coactivator)
Messenger RNA
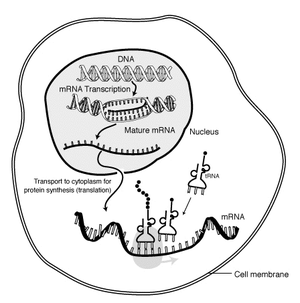
In the ribosome, the nucleic acid polymer is translated into a polymer of amino acids: a protein.
Pre-Initiation
RNA polymerase II binds to the DNA and, along with other cofactors, unwinds the DNA to create an initiation bubble so that the polymerase has access to the single-stranded DNA template. RNA Polymerase II does require a promoter like sequence.
Proximal (core) Promoters: TATA promoters are found around -30 bp to the start site of transcription. Not all genes have TATA box promoters and there exists TATA-less promoters as well. The TATA promoter consensus sequence is TATA(A/T)A(A/T).
The following are the steps involved in TATA Promoter Complex formation:
1. General transcription factors bind
2. TFIID, TFIIA, TFIIB, TFIIF (w/RNA Polymerase II), TFIIH/E
From the beginning of complex formation, the pre-initiation complex is closed.
Initiation

A collection of transcription factors mediate the binding of RNA polymerase II and the initiation of transcription. Only after certain transcription factors are attached to the promoter does the polymerase bind to it. The completed assembly of transcription factors and RNA polymerase II (transcription initiation complex) bind to the promoter.
Once the complex is opened by TFIIH initiation starts and the first phosphodiester bond is formed.
Promoter Clearance
During the formation of the first nucleotide bond the RNA polymerase begins to clear the promoter. There is a tendency to release the RNA transcript and produce truncated transcripts (abortive initiation). Once the transcript reaches approximately 23 nucleotides it no longer slips and Elongation can occur. This is an ATP dependent process. The short-lived, unprocessed or partially processed, product is termed pre-mRNA.
Promoter clearance also coincides with Phosphorylation of serine 5 on the carboxy terminal domain which is phosphorylated by TFIIH.
Eukaryotic pre-mRNA processing
Pre-mRNA requires extensive processing.
5' cap addition
A 5' cap (also termed an RNA cap, an RNA 7-methylguanosine cap or an RNA m7G cap) is a modified guanine nucleotide that is added to the "front" or 5' end of a the pre-mRNA using a 5',5-Triphosphate linkage shortly after the start of transcription. The 5' cap consists of a terminal 7-methylguanosine residue which is linked through a 5'-5'-triphosphate bond to the first transcribed nucleotide. Its presence is critical for recognition and proper attachment of mRNA by the ribosome and protection from 5' exonucleases RNases.
Cap addition is coupled to transcription, and occurs co-transcriptionally, such that each influences the other. Shortly after the start of transcription, the 5' end of the mRNA being synthesized is bound by a cap-synthesizing complex associated with RNA polymerase. This enzymatic complex catalyzes the chemical reactions that are required for mRNA capping. Synthesis proceeds as a multi-step biochemical reaction.
Coding regions
Coding regions are composed of codons, which are decoded and translated into one protein by the ribosome. Coding regions begin with the start codon and end with the a stop codons. Generally, the start codon is an AUG triplet and the stop codon is UAA, UAG, or UGA. The coding regions tend to be stabilised by internal base pairs, this impedes degradation.[39][40] In addition to being protein-coding, portions of coding regions may serve as regulatory sequences in the pre-mRNA as exonic splicing enhancers or exonic splicing silencers.
Untranslated regions
Untranslated regions (UTRs) are sections of the mRNA before the start codon and after the stop codon that are not translated, termed the five prime untranslated region (5' UTR) and three prime untranslated region (3' UTR), respectively. These regions are transcribed with the coding region and thus are exonic as they are present in the mature mRNA. Several roles in gene expression have been attributed to the untranslated regions, including mRNA stability, mRNA localization, and translational efficiency. The ability of a UTR to perform these functions depends on the sequence of the UTR and can differ between mRNAs.
The stability of mRNAs may be controlled by the 5' UTR and/or 3' UTR due to varying affinity for RNA degrading enzymes called ribonucleases and for ancillary proteins that can promote or inhibit RNA degradation.
Translational efficiency, including sometimes the complete inhibition of translation, can be controlled by UTRs. Proteins that bind to either the 3' or 5' UTR may affect translation by influencing the ribosome's ability to bind to the mRNA. MicroRNAs bound to the 3' UTR also may affect translational efficiency or mRNA stability.
Cytoplasmic localization of mRNA is thought to be a function of the 3' UTR. Proteins that are needed in a particular region of the cell can actually be translated there; in such a case, the 3' UTR may contain sequences that allow the transcript to be localized to this region for translation.
Some of the elements contained in untranslated regions form a characteristic secondary structure when transcribed into RNA. These structural mRNA elements are involved in regulating the mRNA. Some, such as the SECIS element, are targets for proteins to bind. One class of mRNA element, the riboswitches, directly bind small molecules, changing their fold to modify levels of transcription or translation. In these cases, the mRNA regulates itself.
Splicing
Splicing is the process by which pre-mRNA is modified to remove certain stretches of non-coding sequences called introns; the stretches that remain include protein-coding sequences and are called exons. Sometimes pre-mRNA messages may be spliced in several different ways, allowing a single gene to encode multiple proteins. This process is called alternative splicing. Splicing is usually performed by an RNA-protein complex called the spliceosome, but some RNA molecules are also capable of catalyzing their own splicing (see ribozymes).
Editing
In some instances, an mRNA will be edited, changing the nucleotide composition of that mRNA. In humans is the apolipoprotein B mRNA, which is edited in some tissues, but not others. The editing creates an early stop codon, which upon translation, produces a shorter protein.
Polyadenylation
Polyadenylation is the covalent linkage of a polyadenylyl moiety to a messenger RNA molecule. Most messenger RNA (mRNA) molecules are polyadenylated at the 3' end. The poly(A) tail and the protein bound to it aid in protecting mRNA from degradation by exonucleases. Polyadenylation is also important for transcription termination, export of the mRNA from the nucleus, and translation. mRNA can also be polyadenylated in prokaryotic organisms, where poly(A) tails act to facilitate, rather than impede, exonucleolytic degradation. The 3' poly(A) tail is a long sequence of adenine nucleotides (often several hundred) at the 3' end of the pre-mRNA. This tail promotes export from the nucleus and translation, and protects the mRNA from degradation.
Monocistronic versus polycistronic mRNA
An mRNA molecule is said to be monocistronic when it contains the genetic information to translate only a single protein. This is the case for most of the eukaryotic mRNAs[41].
Elongation

One strand of DNA, the template strand (or non-coding strand), is used as a template for RNA synthesis. As transcription proceeds, RNA polymerase II traverses the template strand and uses base pairing complementarity with the DNA template to create an RNA copy. Although polymerase traverses the template strand from 3' → 5', the coding (non-template) strand is usually used as the reference point, so transcription is said to go from 5' → 3'. This produces an RNA molecule from 5' → 3', an exact copy of the coding strand (except that thymines are replaced with uracils, and the nucleotides are composed of a ribose (5-carbon) sugar where DNA has deoxyribose (one less oxygen atom) in its sugar-phosphate backbone).
mRNA transcription can involve multiple RNA polymerases on a single DNA template and multiple rounds of transcription (amplification of particular mRNA), so many mRNA molecules can be produced from a single copy of a gene. This step also involves a proofreading mechanism that can replace incorrectly incorporated bases.
The polymerase can experience pauses. These pauses may be intrinsic to RNA polymerase II or due to chromatin structure. Often the polymerase pauses to allow appropriate RNA editing factors to bind.
Termination

Transcription termination in eukaryotes is less well understood, but involves cleavage of the new transcript, followed by template-independent addition of As at its new 3' end, in a process called polyadenylation.
Polyadenylation occurs during and immediately after transcription of DNA into RNA. After transcription has been terminated, the mRNA chain is cleaved through the action of an endonuclease complex associated with RNA polymerase II. After the mRNA has been cleaved, around 250 adenosine residues are added to the free 3' end at the cleavage site. This reaction is catalyzed by polyadenylate polymerase. Just as in alternative splicing, there can be more than one polyadenylation variant of a mRNA.
Once completely processed, the mRNA is termed mature mRNA.
Structure

Transport
Because eukaryotic transcription and translation is compartmentally separated, eukaryotic mRNAs are exported from the nucleus to the cytoplasm. Mature mRNAs are recognized by their processed modifications and then exported through the nuclear pore.
Translation
Eukaryotic mRNA that has been processed and transported to the cytoplasm (i.e. mature mRNA) can then be translated by the ribosome. Translation may occur at ribosomes free-floating in the cytoplasm, or directed to the endoplasmic reticulum by the signal recognition particle.
Post-translational modification
This may include the formation of disulfide bridges or attachment of any of a number of biochemical functional groups, such as acetate, phosphate, various lipids and carbohydrates. Enzymes may also remove one or more amino acids from the leading (amino) end of the polypeptide chain, leaving a protein consisting of two polypeptide chains connected by disulfide bonds.
Protein folding
During and after synthesis, polypeptide chains often fold to assume, so called, native secondary and tertiary structures.
Degradation
After a certain amount of time, the message is degraded by RNases. The limited lifetime of mRNA enables a cell to alter protein synthesis rapidly in response to its changing needs.
Different mRNAs within the same cell have distinct lifetimes (stabilities). In mammalian cells, mRNA lifetimes range from several minutes to days. The greater the stability of an mRNA, the more protein may be produced from that mRNA. The presence of AU-rich elements in some mammalian mRNAs tends to destabilize those transcripts through the action of cellular proteins that bind these motifs. Rapid mRNA degradation via AU-rich elements is a critical mechanism for preventing the overproduction of potent cytokines such as tumor necrosis factor (TNF) and granulocyte-macrophage colony stimulating factor (GM-CSF).[42] Base pairing with a small interfering RNA (siRNA) or microRNA (miRNA) can also accelerate mRNA degradation.
References
- ^ Young VR (1994). "Adult amino acid requirements: The case for a major revision in current recommendations" (PDF). J. Nutr. 124 (8 Suppl): 1517S–1523S. PMID 8064412.
- ^ Fürst P, Stehle P (2004). "What are the essential elements needed for the determination of amino acid requirements in humans?". J. Nutr. 134 (6 Suppl): 1558S–1565S. PMID 15173430.
- ^ Reeds PJ (2000). "Dispensable and indispensable amino acids for humans". J. Nutr. 130 (7): 1835S–40S. PMID 10867060.
- ^ Imura K, Okada A (1998). "Amino acid metabolism in pediatric patients". Nutrition. 14 (1): 143–8. doi:10.1016/S0899-9007(97)00230-X. PMID 9437700.
- ^ "Entrez Gene: APRT adenine phosphoribosyltransferase".
- ^ Robert J. Brooker Genetics: analysis and principles. 2nd edition. (New York: McGraw-Hill 2005) Chapter 12 "Gene transcription and RNA modification" pp. 318-325.
- ^ Latchman DS (1997). "Transcription factors: an overview". Int. J. Biochem. Cell Biol. 29 (12): 1305–12. doi:10.1016/S1357-2725(97)00085-X. PMID 9570129.
- ^ Karin M (1990). "Too many transcription factors: positive and negative interactions". New Biol. 2 (2): 126–31. PMID 2128034.
- ^ Roeder RG (1996). "The role of general initiation factors in transcription by RNA polymerase II". Trends Biochem. Sci. 21 (9): 327–35. doi:10.1016/0968-0004(96)10050-5. PMID 8870495.
- ^ Nikolov DB, Burley SK (1997). "RNA polymerase II transcription initiation: a structural view". Proc. Natl. Acad. Sci. U.S.A. 94 (1): 15–22. doi:10.1073/pnas.94.1.15. PMID 8990153.
- ^ a b Lee TI, Young RA (2000). "Transcription of eukaryotic protein-coding genes". Annu. Rev. Genet. 34: 77–137. doi:10.1146/annurev.genet.34.1.77. PMID 11092823.
- ^ Mitchell PJ, Tjian R (1989). "Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins". Science. 245 (4916): 371–8. doi:10.1126/science.2667136. PMID 2667136.
- ^ Ptashne M, Gann A (1997). "Transcriptional activation by recruitment". Nature. 386 (6625): 569–77. doi:10.1038/386569a0. PMID 9121580.
- ^ Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA (2004). "Structure and evolution of transcriptional regulatory networks". Curr. Opin. Struct. Biol. 14 (3): 283–91. doi:10.1016/j.sbi.2004.05.004. PMID 15193307.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Brivanlou AH, Darnell JE (2002). "Signal transduction and the control of gene expression". Science. 295 (5556): 813–8. doi:10.1126/science.1066355. PMID 11823631.
- ^ Thomas MC, Chiang CM (2006). "The general transcription machinery and general cofactors". Critical reviews in biochemistry and molecular biology. 41 (3): 105–78. PMID 16858867.
- ^ Orphanides G, Lagrange T, Reinberg D (1996). "The general transcription factors of RNA polymerase II". Genes Dev. 10 (21): 2657–83. doi:10.1101/gad.10.21.2657. PMID 8946909.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kornberg RD (2007). "The molecular basis of eukaryotic transcription". Proc. Natl. Acad. Sci. U.S.A. 104 (32): 12955–61. doi:10.1073/pnas.0704138104. PMID 17670940.
- ^ Kronberg, R.D. (1999). "Eukaryotic transcriptional control". Trends Cell Biol. 9 (12): M46–49. doi:10.1016/S0962-8924(99)01679-7. PMID 10611681.
- ^ "Entrez Gene: POLR2A polymerase (RNA) II (DNA directed) polypeptide A, 220kDa".
- ^ "Entrez Gene: POLR2B polymerase (RNA) II (DNA directed) polypeptide B, 140kDa".
- ^ "Entrez Gene: POLR2C polymerase (RNA) II (DNA directed) polypeptide C, 33kDa".
- ^ "Entrez Gene: POLR2D polymerase (RNA) II (DNA directed) polypeptide D".
- ^ "Entrez Gene: POLR2E polymerase (RNA) II (DNA directed) polypeptide E, 25kDa".
- ^ "Entrez Gene: POLR2F polymerase (RNA) II (DNA directed) polypeptide F".
- ^ "Entrez Gene: POLR2G polymerase (RNA) II (DNA directed) polypeptide G".
- ^ "Entrez Gene: POLR2H polymerase (RNA) II (DNA directed) polypeptide H".
- ^ "Entrez Gene: POLR2I polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa".
- ^ "Entrez Gene: POLR2J polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa".
- ^ "Entrez Gene: POLR2J2 DNA directed RNA polymerase II polypeptide J-related gene".
- ^ "Entrez Gene: POLR2J2 DNA directed RNA polymerase II polypeptide J-related gene".
- ^ "Entrez Gene: POLR2J3 DNA directed RNA polymerase II polypeptide J3".
- ^ "Entrez Gene: POLR2J2 DNA directed RNA polymerase II polypeptide J-related gene".
- ^ "Entrez Gene: POLR2J4 DNA directed RNA polymerase II polypeptide J4, pseudogene".
- ^ "Entrez Gene: POLR2J2 DNA directed RNA polymerase II polypeptide J-related gene".
- ^ "Entrez Gene: POLR2K polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa".
- ^ "Entrez Gene: RP11-90M2.3 novel protein similar to polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa POLR2K".
- ^ "Entrez Gene: POLR2L polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa".
- ^ Shabalina SA, Ogurtsov AY, Spiridonov NA (2006). "A periodic pattern of mRNA secondary structure created by the genetic code". Nucleic Acids Res. 34 (8): 2428–37. doi:10.1093/nar/gkl287. PMC 1458515. PMID 16682450.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Katz L, Burge CB (2003). "Widespread selection for local RNA secondary structure in coding regions of bacterial genes". Genome Res. 13 (9): 2042–51. doi:10.1101/gr.1257503. PMC 403678. PMID 12952875.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^
Kozak, M. (1983). "Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles" (PDF). Microbiological Reviews. 47 (1): 1–45. PMID 6343825. Retrieved 2006-08-12.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Shaw G, Kamen R (1986). "A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation". Cell. 46 (5): 659–67. doi:10.1016/0092-8674(86)90341-7. PMID 3488815.
{{cite journal}}: Unknown parameter|month=ignored (help)
Further reading
- Doolittle, R.F. (1989) Redundancies in protein sequences. In Predictions of Protein Structure and the Principles of Protein Conformation (Fasman, G.D. ed) Plenum Press, New York, pp. 599-623
- David L. Nelson and Michael M. Cox, Lehninger Principles of Biochemistry, 3rd edition, 2000, Worth Publishers, ISBN 1-57259-153-6
- Uwe Meierhenrich, Amino acids and the asymmetry of life, Springer-Verlag, 2008, ISBN 978-3-540-76885-2
See also
External links
Amino Acids
- Protein isoelectric point according amino acids charge
- Learn the 20 proteinogenic amino acids online
- The PDF List of Standard Amino Acids
- Molecular Expressions: The Amino Acid Collection - Has detailed information and microscopy photographs of each amino acid.
- Nomenclature and Symbolism for Amino Acids and Peptides International Union of Pure and Applied Chemistry and The International Union of Biochemistry and Molecular Biology. IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN)
- Amino acids overview at Peptide Guide
- Amino acid properties - Properties of the amino acids (a tool aimed mostly at molecular geneticists trying to understand the meaning of mutations)
- Right-handed amino acids were left behind
- Amino acid solutions pH, titration curves and distribution diagrams - freeware
- Synthesis of Amino Acids and Derivatives
- 22nd amino acid - Press release from Ohio State describing the discovery of pyrrolysine as a 22nd amino acid.
- NCBI Bookshelf Free Textbook Access
Nucleotide synthesis
Protein synthesis
- Protein Synthesis
- Protein Synthesis Animation Wesleyan University Learning Objects animation of protein synthesis.
- Science aid:Protein synthesis For high school
Transcription
- Transcription
- Interactive Java simulation of transcription initiation. From Center for Models of Life at the Niels Bohr Institute.
- From RNA to Protein Synthesis hypervideo.
Translation
- Translation of mRNA Section of The Cell: A Molecular Approach by Geoffrey M. Cooper