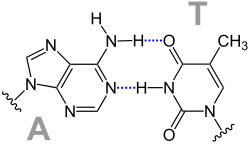
Base pair
As a base pair is referred to in a double strand of a double-stranded nucleic acid ( DNA or RNA , in this case, after English double stranded as dsDNA or dsRNA referred to) two opposing nucleobases which are mutually complementary and are by hydrogen bonds are held together.
While the length of single-stranded nucleic acids (ssRNA or ssDNA, English single stranded ) is given with the number of nucleotides ( nt ) or bases (b) - in the synthetic case sometimes also with the general designation -mer for the number of sequences of a chain molecule, the size of double-stranded DNA sections is usually given in base pairs, abbreviated bp ,
- 1 nt = one nucleotide
- 1 bp = one base pair
- 1 kbp or kb = 1000 base pairs (kilobase pairs)
- 1 Mbp or Mb = 1,000,000 base pairs (mega base pairs)
- 1 Gbp or Gb = 1,000,000,000 base pairs (giga base pairs)
The unduplicated haploid human genome in the nucleus of a germ cell comprises over 3 billion base pairs, about 3.2 Gbp, distributed over 23 chromosomes ( 1 n ; 1 c ). A somatic cell of the human body usually contains a diploid (two times) nuclear chromosomes , or about 6.4 GBP, (2 to 46 chromosomes n ; 2c). This is duplicated (doubled) before a cell division , so that each of the 46 chromosomes consists of two chromatids - identical copies with the same genetic information - before the nuclear division begins as mitosis , with about 13 Gbp (2 n ; 4c). In addition to this nuclear DNA ( nuclear DNA , nDNA), most human cells, as in all eukaryotes, contain a further genome (mitogenome) in each mitochondrion , each about 16.6 kbp ( mitochondrial DNA , mtDNA). An exception are the mature red blood cells , which, like all mammals, have neither a nucleus nor mitochondria. Plant cells also contain the plastid genome (plastome) of their chloroplasts (abbreviated ctDNA or cpDNA).
The number of base pairs is also an important measure of the amount of information that is stored in a gene . Since each base pair represents a choice of 4 possible forms, 1 bp corresponds to the information content of 2 bits , which is twice a bit in the binary code . However, only a small part of the DNA in the human genome carries the genetic information for the construction of proteins; over 95% are non-coding and often consist of repetitive elements .
meaning
Base pairing plays an essential role for DNA reduplication , for transcription and translation in the course of protein biosynthesis, as well as for various designs of the secondary structure and tertiary structure of nucleic acids .
- During the replication , the DNA double strand is separated and the two complementary single strands are supplemented by base pairing from deoxyribonucleotides to form two DNA double strands.
- During transcription , a codogenic strand segment of the DNA is used as a template to build up a single RNA strand with a complementary base sequence by base pairing from ribonucleotides , where A is paired with U. The RNA strands formed serve various purposes as mRNA , tRNA or rRNA .
- During translation , the base sequence of an mRNA section is read in steps of three by pairing the three bases of the anticodon of tRNAs with the complementary base triplets of the mRNA. The base sequence stored in DNA and transcribed in mRNA is translated into a sequence of amino acids with the amino acids transported by tRNA and thus encodes the amino acid sequence as the primary structure of a protein . The wobble pairings also occur when the 3rd base of a codon of the mRNA is paired with the 1st base of the tRNA.
Mating rules
A base pair is formed by a hydrogen bond between two nucleobases . One of the purine bases guanine or adenine is connected to one of the pyrimidine bases cytosine , thymine or uracil to form a pair. In the case of the complementary base pairings between two strand sections of nucleic acids, guanine forms a pair with cytosine, and adenine forms a pair with thymine or uracil. This can result in the following pairings:
DNA / DNA
- Guanine with cytosine: GC or CG
- Adenine with thymine: AT and TA, respectively
DNA / RNA
- Guanine with cytosine: GC or CG
- Adenine with thymine: TA
- Adenine with uracil: AU
RNA / RNA
- Guanine with cytosine: GC or CG
- Adenine with uracil: AU or UA
Watson-Crick pairings
As early as 1949, the Austrian biochemist Erwin Chargaff established with the Chargaff rules that the number of bases adenine (A) and thymine (T) in the DNA is always in a ratio of 1: 1, and the ratio of the bases guanine (G) and cytosine (C) 1: 1. In contrast, the quantitative ratio A: G or C: T varies greatly (Chargaff's rules).
From this, James D. Watson and Francis Harry Compton Crick concluded that AT and GC each form complementary base pairs.
Base pairings also occur in the tRNA and rRNA when the nucleotide strand forms loops and thus complementary base sequences are opposed. Since only uracil is built into the RNA instead of thymine, the pairings are AU and GC.
Unusual pairings
Unusual pairings occur mainly in tRNAs and in triple helices . Although they follow the Watson-Crick scheme, they form other hydrogen bonds: Examples are Reverse-Watson-Crick pairings, Hoogsteen pairings (named after Karst Hoogsteen , born 1923) and Reverse-Hoogsteen pairings
Wobble pairings
The name refers to the wobble hypothesis by Francis Crick (1966). Wobble pairings are the non-Watson-Crick pairings GU or GT and AC:
Web links
- A large database with numerous base pair structures (Department of Biology and Biochemistry, University of Houston )
Individual evidence
- ↑ See dimer , trimer , oligomer , polymer , see for example pegaptanib , a 27mer.
- ^ Hogan, C. Michael (2010). Deoxyribonucleic acid . Encyclopedia of Earth. National Council for Science and the Environment. Eds. S. Draggan, C. Cleveland. Washington DC
- ↑ ctDNA - chloroplast DNA . AllAcronyms.com. Archived from the original on June 3, 2013.
- ^ The Oxford Dictionary of Abbreviations 1998.