Peroxisome
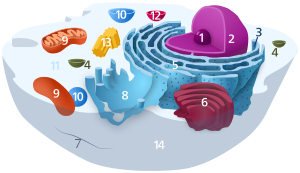
1. Nucleolus (nuclear body)
2. Cell nucleus (nucleus)
3. Ribosomes
4. Vesicle
5. Rough (granular) ER (ergastoplasm)
6. Golgi apparatus
7. Cytoskeleton
8. Smooth (agranular) ER
9 . mitochondria
10. lysosome
11. cytoplasm (with cytosol and cytoskeleton )
12. peroxisomes
13. centrioles
14 cell membrane
| Parent |
| Organelle |
| Subordinate |
|
Membrane lumen matrix protein complexes |
| Gene Ontology |
|---|
| QuickGO |
Peroxisomes , also called microbodies (obsolete), are cell organelles in eukaryotic cells that are surrounded by a biomembrane. They consume oxygen in a variety of metabolic functions and are therefore considered to be the first detoxification devices that became necessary when the earth's atmosphere contained oxygen .
Structural structure

Peroxisomes are small (100-1000 nm diameter) vesicles covered with a simple membrane that are located in the cytoplasm of a cell. In these spatially separated areas ( cell compartments ), protected by the membrane, reactions can take place that would be dangerous for the cell if they were to take place in the cytoplasm. This is an example of the importance of cell compartmentalization. Peroxisomes contain enzymes for the metabolism of hydrogen peroxide (H 2 O 2 ), which is why the term “peroxisome” became established. Morphologically, they used to be referred to as “microbodies”.
The number, size and protein content of the peroxisomes depend on the cell type and growth conditions. For example, it has been observed in baker's yeast ( S. cerevisiae ) that with a good glucose supply only a few, small peroxisomes are present. If, on the other hand, the yeast were supplied with long-chain fatty acids, 20 to 25 large organelles were formed.
Molecular oxygen often serves as a co-substrate from which hydrogen peroxide (H 2 O 2 ) is then formed. The peroxisomes owe their name to the hydrogen peroxide-degrading peroxidase .
Functions
The peroxisomes contain around 60 enzymes called monooxygenases and oxidases , which catalyze the oxidative breakdown of fatty acids , ethanol and other compounds. These enzymes use molecular oxygen as a co-substrate, so that hydrogen peroxide is formed for cell function. Hydrogen peroxide is a cell poison in the cytoplasm and can destroy many important biomolecules.
Hydrogen peroxide can be broken down in two ways. One possibility for detoxification is its immediate conversion by catalase in a disproportionation reaction , whereby water and oxygen are created:
Peroxisomes also have the eponymous peroxidase. The hydrogen peroxide is consumed for its function according to:
Often the enzyme concentrations are so high that they form crystalline aggregates ( nucleoids ).
After Endosymbiontentheorie in the further course of evolution were bacteria (presumably α-proteobacteria ) by the "Urkaryoten-" (precursor of the eukaryotes , presumably Archaeen the Asgard group , see FIG. Eozyten hypothesis cells) was added, which already has a reasonable oxygen utilization apparatus ( citric acid cycle along with respiratory chain possessed) and thus to ATP synthesis by way of oxidative phosphorylation were capable. These were the forerunners of the "modern" mitochondria .
The peroxisomes were not superfluous, but they were integrated into the catabolism (energy gain); The (high-energy) acetyl-CoA became the link . The figure shows an example of how ethanol is used not only to detoxify hydrogen peroxide, but also to be converted into a metabolite (acetyl-CoA) of general importance in catabolism and anabolism (build-up of fatty acids, cholesterol , etc.). Peroxisomes thus contribute to the metabolism of ethanol.
In addition , they catalyze important steps in the biosynthesis of lipids (plasmalogens) in the myelin sheath of nerves (therefore disturbances in their function are often associated with neurological damage). The specific metabolic pathways that run exclusively in peroxisomes are
- the α-oxidation of phytanic acid
- the β-oxidation of very long-chain, polyunsaturated fatty acids
- the biosynthesis of plasmalogens
- the conjugation of cholic acid in the context of bile acid synthesis
Other forms
Glyoxysomes (also called glyoxisomes ) are specialized peroxisomes that are found in the endosperm and the storage tissues of fatty sperm cells. They got their name because they are involved in the glyoxylate cycle . The enzymes contained in the glyoxysomes enable the use of fats to build up biopolymers ( sugar , proteins ), which are necessary for plant growth.
In photosynthetically active plants, peroxisomes also take part in photorespiration - there also in cooperation with mitochondria. They are known as leaf peroxisomes . Vegetable glyoxysomes and leaf peroxisomes can transform into one another.
Emergence
The origin of the peroxisomes has been the subject of controversy in recent years. Nowadays it is known that peroxisomes, analogous to mitochondria , can multiply by division within the cell. The de novo formation of new peroxisomes is a multi-step process that begins with the constriction of precursor vesicles from the endoplasmic reticulum (ER). The small precursor veins probably then fuse to form a mature peroxisome. Pex3, an integral membrane protein, is essential for biogenesis in yeast. The breakdown of peroxysomes is called peroxyphagia , in analogy to mitophagy (breakdown of mitochondria) and reticulophagy (breakdown of the ER).
Protein transport
Since peroxisomes do not contain ribosomes , all enzymes must be synthesized in the cytosol and then transported into the peroxisome. Here, proteins are brought into the peroxisome post-translationally in the folded state. There are two known ways. Most proteins require a C-terminal signal sequence , the so-called peroxisome targeting signal (peroxisome target signal) PTS1. This signal sequence is shorter than that of proteins that are to be brought into the mitochondrion or the ER; most of the time it consists only of the three amino acids serine-lysine-leucine (SKL). The signal sequence of those “PTS1 proteins” is recognized in the cytosol by Pex5p and carried to the peroxisome, where they are transported into the interior of the peroxisome by a protein membrane complex. The protein-Pex5p complex docks onto the integral membrane protein Pex14. The complex of Pex5 and the protein is then transported into the peroxisome, where Pex5 is split off and is recycled again via the Pex2 / 10/12 membrane complex with consumption of ATP .
In the second transport route, an N-terminal and also longer signal peptide is brought to the protein membrane complex of the peroxisome by Pex7p. This signal sequence is also referred to as PTS2, which means that transported proteins are PTS2 proteins. In addition to Pex7p, a spliced form of Pex5p is also used in mammalian cells. After transport into the matrix of the peroxisome, the signal peptide is then cut off.
Diseases
Diseases in which peroxisomes play a role:
1. Peroxisome defects
- Zellweger Syndrome
- Rhizomele Chondrodysplasia punctata type 1 ( mutation of the PEX7 gene )
- Neonatal adrenoleucodystrophy
- Infantile Refsum Syndrome
2. Peroxisomal enzyme defect
- Pseudo-Zellweger syndrome (mutation of acyl-CoA oxidase)
- X-linked adrenoleukodystrophy (secondary due to peroxisomal transporter protein defect for VLCFA-CoA synthetase)
- Rhizomele Chrondrodysplasia punctata type 2 (mutation of the DHAPAT gene)
See also
literature
- B. Alberts et al. : Molecular Biology of the Cell . Garland Science, 4th edition, 2002. ISBN 0815340729 .
- N. Campbell et al. : Biology . 1st edition, 1st corrected reprint, Spektrum Akademischer Verlag 1997, Heidelberg. ISBN 3-8274-0032-5 .
Individual evidence
- ^ Peter Karlson, Detlef Doenecke, Jan Koolman, Georg Fuchs and Wolfgang Gerok: Karlsons Biochemie und Pathobiochemie . Georg Thieme, 15th edition 2005, ISBN 978-3133578158 ; P. 396f.
- ↑ a b c Peter H. Raven, Ray F. Evert, Susan E. Eichhorn: Biology of plants . 4th edition. Gruyter, Berlin, New York 2006; ISBN 978-3-11-018531-7 ; P. 53f.
- ^ Horst Feldmann: Yeast: Molecular and Cell Biology . Wiley-VCH Verlag GmbH & Co. KGaA 2009; ISBN 978-3527326099 ; P. 159
- ↑ D'Eustachio / reactome: Peroxisomal lipid metabolism ( page no longer available , search in web archives ) Info: The link was automatically marked as defective. Please check the link according to the instructions and then remove this notice.
- ^ Margit Pavelka (eds.) And Jürgen Roth (ed.): Functional Ultrastructure: Atlas of Tissue Biology and Pathology . Springer, Vienna; 2nd edition 2010; ISBN 978-3211993897 ; P. 134
- ↑ Daniel J. Klionsky et al. (2007): How shall I eat thee ? In: Autophagy 3 (5); Pp. 413-416; PMID 17568180 ; PDF (free full text access, English).
- ↑ Lynne Cassimeris, George Plopper and Vishwanath R. Lingappa: Lewin's Cells . Jones & Bartlett Pub (Ma); 2nd edition 2010; ISBN 978-0763766641 ; P. 338
- ↑ Marc Fransen, Stanley R. Terlecky, and Suresh Subramani: Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import , PMC 20933 (free full text)
- ↑ Harvey Lodish: Molecular Cell Biology (Seventh Edition, 2012) pp. 612f. ISBN 978-1464109812


