Translation (biology)
In biology, translation is the term used to describe the synthesis of proteins in the cells of living organisms, which takes place on the ribosomes according to genetic information (see also protein biosynthesis ).
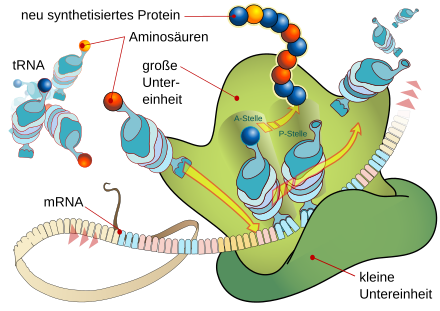
A protein is synthesized on the ribosome, which is composed of its two subunits . Its peptide chain is gradually built up from certain amino acids that are each carried by tRNA molecules. These are only added if the anticodon of a tRNA matches the next codon of the mRNA strand in a base-pairing manner. In this way, the amino acid that is bound to the peptide at the P site is determined in each case at the A site. In this way, the information contained in the base sequence of the mRNA is translated into the encoded formation of the amino acid sequence of a protein.
Translation is an essential part of the gene expression process following transcription , in which the information from a DNA segment is overwritten on individual RNA strands. According to the given information, translation then takes place on the ribosomes in the cytoplasm of a cell. The base sequence of an mRNA molecule is translated into the coded amino acid sequence of a polypeptide and a protein is formed.
General procedure

the base triplet of a codon of the mRNA strand, which consists of three nucleobases , binds the anticodon on the anticodon loop of a tRNA , which is loaded with a certain amino acid at the end of its acceptor arm . This is ready for the next step of the ribosomal synthesis and is linked to the amino acid chain by a peptide bond .
In the genome of every organism , sections can be found which, as genes, contain information not only for the construction of RNA , but also for the construction of proteins . The mRNA ("m" stands for messenger , messenger), which is formed and possibly processed according to the base sequence of such a section of DNA , contains the sequence of its bases , the base sequence , in each case selected information for the biosynthesis of certain proteins.
This genetic information is used in the course of translation as an instruction to synthesize the corresponding protein by translating sections of the base sequence into the amino acid sequence of a peptide according to the genetic code . Three consecutive nucleotides of the mRNA each represent a codon and thus code as a base triplet for a specific amino acid. From the encoded amino acids, the polypeptide chain of a protein is built up sequentially on the ribosome in the order specified by the nucleotide sequence , with the specified amino acid sequence . The information in the mRNA is read here in the 5 '→ 3' direction, i.e. the same direction in which the RNA was also transcribed (by RNA polymerase ) .
Various tRNA molecules are required as amino acid "transporters" for the translation process ("t" stands for transfer ). These can each bind with one of their loops, the anticodon loop , via their anticodon, in a complementary base-pairing manner, to a codon on the mRNA and at their other end, the acceptor arm , are loaded with the amino acid matching the codon through the different aminoacyl-tRNA synthetases .
During translation, the ribosome attaches itself to the mRNA strand and brings it together with a loaded tRNA in such a way that the base triplet of an anticodon of the tRNA can now attach to a base triplet of a codon on the mRNA as a matching counterpart. The actual translation process begins at the point in the mRNA where the base sequence (e.g. → ..., adenine, uracil, guanine, ...) represents the start codon (mostly AUG). A second tRNA, which corresponds to the following codon and which also carries an amino acid, attaches itself to the mRNA next to the first tRNA. The two amino acids positioned next to one another are then linked by a peptide bond, and the first tRNA leaves the ribosome unloaded without an amino acid. A third loaded tRNA that matches the next codon is now attached to the mRNA. Their amino acid is linked to the existing amino acid chain and thus extends it by another link. This process continues, so that an ever longer chain of amino acids is formed. The ribosome that catalyzes this process moves step by step by a triplet or codon on the mRNA. The translation ends when a base triplet is found in this reading frame on the mRNA, which represents a stop codon (e.g. UGA). Usually none of the types of tRNA molecules present can bind to this. The area on an mRNA between the start and the associated stop codon is also called open reading frames ( open reading frame referred to).
At the end of translation, the peptide synthesized as a chain of amino acids detaches from the ribosome and the nascent polypeptide chain folds in the medium to form the native protein, usually in such a way that a complex spatial structure is created ( secondary structure and tertiary structure ). It may combine with other proteins to form higher-level quaternary structures .
An mRNA is usually read several times until it is broken down into its building blocks, the ribonucleotides , by the activity of nucleases . In eukaryotes , the shelf life is increased by post-transcriptional modifications in the nucleus.
Biochemical process
Although there are 61 codons for the 20 canonical proteinogenic amino acids, not as many different types of tRNA are used in the cytoplasm of a cell . In fact, 31 different anticodons are sufficient as mediators between the 20 amino acids and the 61 codons in bacteria. The roughly 600 tRNA genes found in humans represent only 48 different anticodons. Because some tRNAs can recognize several different codons for the same amino acid. This is the case, for example, when the first two bases of a base triplet define a certain amino acid and the third no longer plays a role. The anticodon of the tRNA loaded with the corresponding amino acid primarily recognizes the first two positions of the triplet on the mRNA with the usual complementary base pairing - the third pairing can be wobbly (see also wobble hypothesis ) - and thus various similar codons. In contrast, the tryptophan-loadable tRNA (tRNA Trp ) normally only recognizes a certain codon ( UGG ).
All mature tRNA molecules consist of an RNA strand with a little less than 100 nucleotides, form a clover-like shape in their secondary structure as a result of intramolecular pairings of complementary nucleotide sequences with loops and fold three-dimensionally into a hook-like tertiary structure. The 5 'and 3' ends are united in the so-called acceptor arm. The corresponding amino acid then binds at the 3 'end via a post-transcriptionally added CCA triplet. The anticodon loop lies opposite the acceptor stem in the secondary structure, and it is also the greatest distance apart in the tertiary structure. Three central bases of this loop in the anticodon arm form the anticodon - mostly in position number 36, 35 and 34, the latter then pairing with the 3rd base of the codon. The D loop contains the unusual dihydrouridine (D), the T loop, in addition to thymidine (T), typically contains pseudouridine (Ψ) and cytosine (C). The V-loop is variable, i.e. it is composed differently for individual tRNA types.
A special aminoacyl-tRNA synthetase is responsible for loading a tRNA with its amino acid . Usually there is a specific synthetase for each amino acid.
Ribosomes and Protein Synthesis
On the ribosomes , an aminoacyl-tRNA is paired with the codon of the mRNA via its anticodon, and the polypeptide chain of proteins is synthesized through peptide binding of the amino acids brought in. This ribosomal peptide synthesis through translation of the genetically encoded information is the main step in protein biosynthesis .
Ribosomes consist of two subunits, each made up of RNA ( ribosomal RNA ) and polypeptides (ribosomal proteins). First of all, the two subunits are separate. During translation, they combine and form two areas where the tRNAs can attach: the aminoacyl site (A site) for the tRNA with the next amino acid to be added, the peptidyl site (P site) for the tRNA amino acid added to the growing peptide chain. The discharged tRNA molecules then leave the ribosome via another region, the exit site (E site).
Initiation of translation in prokaryotes
In addition to the two ribosomal subunits and the mRNA, the cell also needs a special tRNA for initiation as the start of the process and the beginning of the chain. This initiator tRNA binds to the start codon AUG and in bacteria is a tRNA i fMet that transmits formylmethionine (fMet) instead of the methionine of the tRNA i Met common in archaea (and eukaryotes) . In addition, three initiation factors (IF 1, IF 2, IF 3) play a role in prokaryotes .
The small subunit (30S) initially forms a complex with the initiation factors 1 and 3. The task of IF1 is to dissociate the non-initiator tRNA (which is in dynamic equilibrium). The IF3, together with the IF1, prevents the two ribosomal subunits from binding prematurely.
IF2, a G protein , binds GTP, undergoes a conformational change and can thus bind the initiator tRNA. This complex of IF2-GTP and (loaded) fMet-tRNA i fMet now has the possibility of binding both to mRNA and to the 30S unit.
The small subunit is able to recognize the appropriate binding site through an interaction of the anti-Shine-Dalgarno sequence of its 16S rRNA ( ribosomal RNA as part of the 30S unit) with the Shine-Dalgarno sequence on the mRNA. This non-coding sequence lies a few nucleotides (9 nt upstream) in front of a base triplet, which represents an AUG , and thus enables the start codon to be recognized by the initiator tRNA. The completion of the initiation is initiated by GTP hydrolysis at the IF2. The initiation factors are released and only then do the 50S subunit bind, creating the 70S initiator complex. The fMet-tRNA i fMet is already in the P site of the 50S subunit at the beginning of translation. The other two digits, A and E, are empty.
Elongation of the polypeptide chain
Elongation is the process of lengthening the chain of amino acids; it takes place at the location of the ribosome's recognition and binding. A single elongation step contains three steps: binding of the loaded tRNA, formation of the peptide bond and preparation for the next elongation step. This is repeated until a terminating codon is reached.
Termination in prokaryotes
The end of translation is reached when one of the stop triplets UAG, UAA or UGA appears in the A-site of the ribosome. Since there is no matching tRNA for these codons in the cell, translation stops.
Termination factors (release factors) then bind to the base triplet of the stop codon: RF1 to UAG and UAA or RF2 to UAA and UGA. This causes the bond between the last amino acid and the last tRNA in the ribosome to be broken. During translation, the ester cannot be broken up by hydrolysis, since the peptidyl transferase region is completely anhydrous. This prevents spontaneous hydrolysis during elongation. The RF brings exactly one molecule of water into the peptidyl transferase center, mediated by the amino acid sequence glycine-glycine-glutamine. This can then cleave the ester bond with the help of the catalytic activity of the ribosome. This sequence is also found in the eukaryotic RF. The dissociation of RF1 / RF2 from the ribosome is catalyzed by the termination factor RF3.
Now the protein and the mRNA fall off the ribosome, which breaks down again into its two subunits. The initiation factor IF3 maintains the dissociated state. So the cycle can begin again.
Translation in eukaryotes
initiation
Translation in eukaryotes differs from prokaryotic translation in particular in initiation, in which a number of special eukaryotic initiation factors (eIF) are involved. The initiator tRNA here is a tRNA i Met that carries methionine and is not formylated . A Shine-Dalgarno sequence cannot be found on the eukaryotic mRNA. The first base triplet AUG of the mRNA from the 5 'end is usually chosen as the start codon. The 40S subunit is mostly bound to the 5 'cap structure of the mRNA. After the preinitiatin complex has been formed from a small subunit and initiator tRNA with eIF-2 and other factors, the mRNA is searched in the 3 'direction for an AUG. If this search is successful, the initiator Met-tRNA is attached to the base triplet of the mRNA. However, the translation process only begins when the larger subunit (60S) of the ribosome has also been bound (see adjacent figure).
If eukaryotic mRNA form complex secondary structures during processing or their transport from the nucleus, these can be broken up again by helicases .
Termination
The end of translation is usually marked by the base triplet of a stop codon. In the meantime, however, some genes have also been discovered in humans, in which elongated proteins and thus new isoforms are created by reading a stop signal on the mRNA (English called translational readthrough ) . This can happen if, for example, the UGA codon is interpreted differently and translated into an amino acid, such as tryptophan. This should be distinguished from those special cases of recoding in which the use of specific tRNA molecules enables the incorporation of additional proteinogenic amino acids such as selenocysteine and pyrrolysine .
regulation
Every protein the cell needs for survival is encoded in the genes . However, the required amount is not coded directly in the gene and also depends on environmental conditions, age and cell cycle and, above all, on the type of cell (cell type). In quantitative terms, by far the most important point of attack for controlling protein production (protein expression) is not translation, but transcription . The question of whether a certain protein is produced is therefore not primarily decided on whether the mRNA that codes for this protein takes part in translation, but on whether the mRNA is produced at all.
Nevertheless, the regulation of translation is an important point of attack in gene regulation. This controls how much or which protein is produced from a specific mRNA. Two examples:
- Regulation of initiation: The eukaryotic initiation factor eIF2 can be regulated by phosphorylation . The regulation of translation is linked to cell growth or the cell cycle and the amount of available nutrients via the mTOR signal path .
- Regulation of termination: The peroxisomal isoforms of LDH can be produced in the cell through functional translational read-through .
Further key words for the regulation of translation are 5'-positioned small open reading frames, codon optimality , and the start codon context Kozak sequence .
Example of a regulation of translation of ribosomal proteins
Correct expression of ribosomal proteins poses an interesting regulatory problem for the cell. Each ribosome contains around 50 special proteins, all of which have to be synthesized at the same rate. Furthermore, the rate of protein synthesis in the cell and the need for ribosomes are closely related to cell growth. A change in the growth conditions quickly leads to an increase or decrease in the rate of synthesis of these ribosomal components. Regulation is required for this.
The control of the genes for the ribosomal proteins is simplified by the organization in different operons , each of which contains genes for up to 11 ribosomal proteins.
The primary control occurs at the level of translation. This can be demonstrated by the following experiment :
If additional copies of such an operon are introduced into the genome of a cell through genetic engineering , the amount of mRNA generated by transcription increases accordingly. Nevertheless, the rate of synthesis of the protein remains almost unchanged. The cell thus compensates for the increased amount of mRNA. Ribosomal proteins act as repressors of their own translation.
For each operon, an already synthesized ribosomal protein can bind to the mRNA of the operon. This binding site is near one of the operon's first genes. This prevents ribosomes from binding to the mRNA and starting translation. The repression of the translation of the first genes thus prevents the expression of part or all of the remainder of the subsequent genes.
This mechanism is very delicate. For example, just a few molecules of the L4 protein that are not used to form ribosomes prevent the synthesis of this protein as well as the other 10 ribosomal proteins in the same operon. This ensures that the proteins are not produced in too large quantities that cannot be completely used up to form ribosomes.
How a protein can serve both as a ribosomal component and as a regulator of its own translation was investigated by comparing the binding sites of the protein on the rRNA with the binding sites with its own mRNA. Both binding sites are similar in their sequence and their secondary structure . Since the binding of the ribosomal proteins to the rRNA is stronger than that to the mRNA, translation is only suppressed when the need for proteins for the production of ribosomes is met.
Translocation in and through membranes
In both prokaryotes and eukaryotes , protein synthesis takes place on the ribosomes in the cytosol of the cell. From here, proteins can be transported into or through a membrane . This shift to another location, also known as translocation , can already be initiated during the synthesis of a protein during translation, i.e. it can take place cotranslationally , or it can only take place after the synthesis has been completed, i.e. post-translationally .
For the translocation, the preferred mode of transport and the respective destination are mostly certain sections in the amino acid sequence of the protein formed, which are recognized as signal sequences by signal recognition particles or special protein complexes (such as the Sec system ). In prokaryotes, a newly formed protein can be determined for transport into the cell membrane or through it into the extraplasmic space, for example for the construction of a cell wall . Since eukaryotes have different organelles as membrane-covered cell compartments , the possible target locations for a translocation of proteins are more diverse here. The transport routes to target compartments such as the endoplasmic reticulum , cell nucleus , peroxisomes and other vesicles as well as those in mitochondria , chloroplasts or other plastids must be distinguished from transport into the extracellular space or into the cytomembrane .
Cotranslational protein transport
In this way, the ribosome is brought to the cell membrane (especially the endoplasmic reticulum) by signal proteins. The resulting protein is then pushed through a special tunnel into the area behind.
Post-translational protein transport
The protein, which is fully assembled in the cell and protected from premature unfolding by a chaperone , is transported to its destination. In the case of bacteria, a built-in "kink" in the protein makes threading through the cell membrane easier. The eukaryotic post-translational transport across the ER membrane has been demonstrated in yeast.
literature
- Rolf Knippers: Molecular Genetics . 9th completely revised edition. Thieme, Stuttgart et al. 2006, ISBN 3-13-477009-1 .
Web links
- Lukas Hensel - animation for translation
- University of Bern - Translation in prokaryotes and eukaryotes
- Central for teaching media on the Internet - translation
- IUBMB: Prokaryotic and eukaryotic translation factors
- MRC-LabMB: Movies and Overview Figures of the Ribosome
Individual evidence
- ↑ B. Alberts, A. Johnson, J. Lewis et al .: Molecular Biology of the Cell. 4th edition. Garland Science, New York 2002, Chapter: From RNA to Protein. Online on the NCBI bookshelf
- ↑ see entries for Homo sapiens in the genomic tRNA database (GtRNAdb) .
- ↑ James D. Watson et al: Molecular Biology of the Gene . 5th edition. Pearson / Cummings et al., San Francisco CA 2004, ISBN 0-8053-4635-X , pp. 435 .
- ^ F. Schueren and S. Thoms: Functional Translational Readthrough: A Systems Biology Perspective . In: PLOS Genetics . 12 (8), No. e1006196, August 2016, p. 12. doi : 10.1371 / journal.pgen.100619 . PMID 27490485 . PMC 4973966 (free full text).



