Nodaviridae: Difference between revisions
Velayinosu (talk | contribs) |
No edit summary |
||
| Line 11: | Line 11: | ||
}} |
}} |
||
'''''Nodaviridae''''' is a family of [[virus]] |
'''''Nodaviridae''''' is a family of [[nonenveloped]] [[Positive-strand RNA virus|positive-strand RNA viruses]].<ref>{{cite journal |last1=Sahul Hameed |first1=AS |last2=Ninawe |first2=AS |last3=Nakai |first3=T |last4=Chi |first4=SC |last5=Johnson |first5=KL |last6=ICTV Report |first6=Consortium |title=ICTV Virus Taxonomy Profile: Nodaviridae. |journal=The Journal of General Virology |date=January 2019 |volume=100 |issue=1 |pages=3-4 |doi=10.1099/jgv.0.001170 |pmid=30431412|doi-access=free }}</ref> Vertebrates and invertebrates serve as natural hosts. Diseases associated with this family include: viral [[encephalopathy]] and [[retinopathy]] in fishes.There are nine species in the family divided among two genera.<ref name= ICTV>{{cite web |title=ICTV Report Nodaviridae |url=http://www.ictv.global/report/nodaviridae}}</ref><ref name=ViralZone>{{cite web|title=Viral Zone|url=http://viralzone.expasy.org/all_by_species/47.html|publisher=ExPASy|accessdate=15 June 2015}}</ref> |
||
== |
==Structure== |
||
The virus is not enveloped and has an icosahedral capsid (triangulation number=3) ranging from 29 to 35 nm in diameter. The [[capsid]] is constructed of 32 capsomers.<ref name=ICTV/> |
The virus is not enveloped and has an icosahedral capsid (triangulation number=3) ranging from 29 to 35 nm in diameter. The [[capsid]] is constructed of 32 capsomers.<ref name=ICTV/> |
||
== Genome == |
|||
The genome is linear, positive sense, bipartite (composed of two segments—RNA1 and RNA2) single stranded [[RNA]] consisting of 4500 [[nucleotide]]s with a 5’ terminal [[Methylation|methylated]] cap and a non-polyadenylated 3’ terminal.<ref name=ICTV/> |
The genome is linear, positive sense, bipartite (composed of two segments—RNA1 and RNA2) single stranded [[RNA]] consisting of 4500 [[nucleotide]]s with a 5’ terminal [[Methylation|methylated]] cap and a non-polyadenylated 3’ terminal.<ref name=ICTV/> |
||
RNA1, which is ~3.1 kilobases in length, encodes a protein that has multiple functional domains: a mitochondrial targeting domain, a transmembrane domain, an RNA-dependent RNA polymerase (RdRp) domain, a self-interaction domain and an RNA capping domain. In addition, RNA1 encodes a subgenomic RNA3 that encodes protein B2, an RNA silencing inhibitor.<ref name=ICTV/> |
RNA1, which is ~3.1 kilobases in length, encodes a protein that has multiple functional domains: a mitochondrial targeting domain, a transmembrane domain, an RNA-dependent RNA polymerase (RdRp) domain, a self-interaction domain and an RNA capping domain. In addition, RNA1 encodes a subgenomic RNA3 that encodes protein B2, an RNA silencing inhibitor.<ref name=ICTV/> |
||
| ⚫ | |||
{| class="wikitable sortable" style="text-align:center" |
|||
|- |
|||
! Genus !! Structure || Symmetry !! Capsid !! Genomic arrangement !! Genomic segmentation |
|||
|- |
|||
|Betanodavirus||Icosahedral||T=3||Non-enveloped||Linear||Segmented |
|||
|- |
|||
|Alphanodavirus||Icosahedral||T=3||Non-enveloped||Linear||Segmented |
|||
|} |
|||
| ⚫ | |||
==Life cycle== |
==Life cycle== |
||
Viral replication is cytoplasmic. Entry into the host cell is achieved by penetration into the host cell. Replication follows the positive stranded RNA virus replication model. Positive stranded RNA virus transcription, using the internal initiation model of subgenomic RNA transcription is the method of transcription. Vertebrates and invertebrates serve as the natural host. Transmission routes are contact and contamination.<ref name=ICTV/><ref name=ViralZone /> |
Viral replication is cytoplasmic. Entry into the host cell is achieved by penetration into the host cell. Replication follows the positive stranded RNA virus replication model. Positive stranded RNA virus transcription, using the internal initiation model of subgenomic RNA transcription is the method of transcription. Vertebrates and invertebrates serve as the natural host. Transmission routes are contact and contamination.<ref name=ICTV/><ref name=ViralZone /> |
||
| Line 41: | Line 34: | ||
==Taxonomy== |
==Taxonomy== |
||
The members of the genus ''Alphanodavirus'' were originally isolated from insects while those of the genus ''Betanodavirus'' were isolated from fish. A small number of nodoviruses seem to lie outside either of these clades.<ref name=ICTV/> |
The members of the genus ''Alphanodavirus'' were originally isolated from insects while those of the genus ''Betanodavirus'' were isolated from fish. A small number of nodoviruses seem to lie outside either of these clades.<ref name=ICTV/> While NoV remains the type species of ''Alphanodavirus'', ''[[Flock house virus]]'' (FHV) is the best studied of the nodaviruses.<ref name="ICTV" /> There are nine species in this family divided among two genera:<ref name="ICTV" /> |
||
While NoV remains the type species of ''Alphanodavirus'', ''[[Flock house virus]]'' (FHV) is the best studied of the nodaviruses.<ref name=ICTV /> |
|||
==History== |
==History== |
||
Revision as of 10:07, 1 December 2020
| Nodaviridae | |
|---|---|
| |
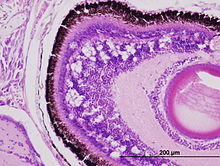
| Vacuoles in retina of Australian bass larva experimentally infected with Betanodavirus | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Kitrinoviricota |
| Class: | Magsaviricetes |
| Order: | Nodamuvirales |
| Family: | Nodaviridae |
| Genera | |
Nodaviridae is a family of nonenveloped positive-strand RNA viruses.[1] Vertebrates and invertebrates serve as natural hosts. Diseases associated with this family include: viral encephalopathy and retinopathy in fishes.There are nine species in the family divided among two genera.[2][3]
Structure
The virus is not enveloped and has an icosahedral capsid (triangulation number=3) ranging from 29 to 35 nm in diameter. The capsid is constructed of 32 capsomers.[2]
Genome
The genome is linear, positive sense, bipartite (composed of two segments—RNA1 and RNA2) single stranded RNA consisting of 4500 nucleotides with a 5’ terminal methylated cap and a non-polyadenylated 3’ terminal.[2]
RNA1, which is ~3.1 kilobases in length, encodes a protein that has multiple functional domains: a mitochondrial targeting domain, a transmembrane domain, an RNA-dependent RNA polymerase (RdRp) domain, a self-interaction domain and an RNA capping domain. In addition, RNA1 encodes a subgenomic RNA3 that encodes protein B2, an RNA silencing inhibitor.[2]
RNA2 encodes protein α, a viral capsid protein precursor, which is auto-cleaved into two mature proteins, a 38 kDa β protein and a 5 kDa γ protein, at a conserved Asn/Ala site during virus assembly.[2]
Life cycle
Viral replication is cytoplasmic. Entry into the host cell is achieved by penetration into the host cell. Replication follows the positive stranded RNA virus replication model. Positive stranded RNA virus transcription, using the internal initiation model of subgenomic RNA transcription is the method of transcription. Vertebrates and invertebrates serve as the natural host. Transmission routes are contact and contamination.[2][3]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Betanodavirus | Fish | None | Unknown | Lysis | Cytoplasm | Cytoplasm | Passive diffusion, direct contact |
| Alphanodavirus | Insects, mammals, fishes | None | Unknown | Lysis | Cytoplasm | Cytoplasm | Unknown |
Taxonomy
The members of the genus Alphanodavirus were originally isolated from insects while those of the genus Betanodavirus were isolated from fish. A small number of nodoviruses seem to lie outside either of these clades.[2] While NoV remains the type species of Alphanodavirus, Flock house virus (FHV) is the best studied of the nodaviruses.[2] There are nine species in this family divided among two genera:[2]
History
The name of the family is derived from the Japanese village of Nodamura, Iwate Prefecture where Nodamura virus was first isolated from Culex tritaeniorhynchus mosquitoes.
References
- ^ Sahul Hameed, AS; Ninawe, AS; Nakai, T; Chi, SC; Johnson, KL; ICTV Report, Consortium (January 2019). "ICTV Virus Taxonomy Profile: Nodaviridae". The Journal of General Virology. 100 (1): 3–4. doi:10.1099/jgv.0.001170. PMID 30431412.
- ^ a b c d e f g h i "ICTV Report Nodaviridae".
- ^ a b "Viral Zone". ExPASy. Retrieved 15 June 2015.
