Coronaviridae
| Coronaviridae | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|

Coronaviridae |
||||||||||||||||
| Systematics | ||||||||||||||||
|
||||||||||||||||
| Taxonomic characteristics | ||||||||||||||||
|
||||||||||||||||
| Scientific name | ||||||||||||||||
| Coronaviridae | ||||||||||||||||
| Left | ||||||||||||||||
|
Coronaviridae is a virus family within the order Nidovirales . The viruses within the family are colloquially called coronaviruses and are among the RNA viruses with the largest genomes .
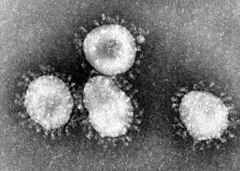
The first coronaviruses were described as early as the mid-1960s. The British virologist June Almeida is considered to be the discoverer and was able to take a picture using an electron microscope in 1966. The roughly spherical viruses in the electron microscope image are conspicuous by a wreath of petal-like appendages that are reminiscent of a solar corona and that gave them their name.
Representatives of this virus family cause very different diseases in all four classes of terrestrial vertebrates ( mammals , birds , reptiles , amphibians ). They are genetically highly variable and so can sometimes infect several types of hosts . In humans, seven types of coronavirus are important as pathogens from mild respiratory infections (colds / flu-like infections) to so-called severe acute respiratory syndrome (SARS).
The following coronaviruses have become particularly well-known among the human coronaviruses:
- SARS-CoV [-1] (severe acute respiratory syndrome coronavirus [1])
- MERS-CoV (Middle East respiratory syndrome coronavirus)
- SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2)
They were or are the triggers of the SARS pandemic 2002/2003 , the MERS epidemic (from 2012) and the COVID-19 pandemic (from 2019).
Note: For a clearer and more meaningful distinction between the coronaviruses SARS-CoV and SARS-CoV-2, SARS-CoV is sometimes also referred to as SARS-CoV-1.
features
Appearance
The 60 to 160 nm large virus particles ( virions ) have a virus envelope in which several different types of membrane proteins are embedded. The characteristic appearance of the coronavirus ( Latin corona , crown, crown ' ) is due to many club-shaped structures on the surface that protrude about 20 nm outward, the peplomeres called spikes . They consist of parts of the large glycosylated S protein (spikes protein, 180 to 220 kDa ), which here forms a membrane-anchored trimer . These parts carry both (S1) the receptor binding domain (RBD), with which the virus can dock onto a cell, and (S2) a subunit which, as a fusion protein (FP), causes the virus envelope and cell membrane to fuse .
The smaller envelope protein (E protein, 9 to 12 kDa) is present in smaller quantities on the outside , only in the HCoV-OC43 ( human coronavirus OC43 ) and the coronaviruses of group 2 (genus Betacoronavirus ) is the hemagglutin Esterase protein (HE protein, 65 kDa). The M protein (matrix protein, 23 to 35 kDa), which is also anchored in the membrane envelope, is directed inwards and a matrix protein on the inside of the virus envelope.
Inside the shell there is a presumably icosahedral capsid that contains a helical nucleoprotein complex . This consists of the nucleoprotein N (50 to 60 kDa), which is complexed with the strand of a single-stranded RNA of positive polarity . Certain amino acid residues of the N protein interact with the matrix protein M, so that the capsid is associated with the inside of the membrane.
Genome
The single-stranded RNA genome of the coronaviruses is about 27,600 to 31,000 nucleotides (nt) long, which means that coronaviruses have the longest genomes of all known RNA viruses .
At the 5 'end there is a 5' cap structure and a non-coding region ( English untranslated region , UTR) of about 200 to 400 nt, which contains a 65 to 98 nt short, so-called leader sequence . At the 3 'end of a further adds UTR of 200 to 500 nt, in a poly (A) tail ends. The genome of coronaviruses containing 6 to 14 Open reading frames ( English open reading frames , ORFs), of which the two largest (the genes for the non-structural proteins NSP-1a and 1b), close to lie at the 5 'end and with different reading frames slightly overlap . The point of overlap forms a hairpin structure , which enables a frameshift during translation on the ribosomes in 20 to 30% of the reading runs and thus leads to the synthesis of smaller amounts of NSP-1b.
In addition to replicating their genome, the viruses synthesize (depending on the virus species) 4–9 mRNA molecules, the 5 'and 3' ends of which are identical to those of the genome. These "nested" mRNAs are also known as "nested set of mRNAs" and have contributed to the naming of the superordinate virus order, nidovirales (from the Latin nidus 'nest' ).
In contrast to the usually high error rate of the RNA polymerase of other RNA viruses, which leads to a restriction of the genome length to about 10,000 nucleotides, coronaviruses have a relatively high genetic stability (preservation), among other things through a 3'-5'- exoribonuclease - Function of the protein NSP-14 achieved. Presumably, this proof-reading mechanism prevents the antiviral agent ribavirin from working in COVID-19 (SARS-CoV-2).
Occurrence and distribution
Infectious bronchitis in poultry caused by the infectious bronchitis virus (IBV, type Avian coronavirus ), a coronavirus, was examined as early as 1932 . At that time, the examinations concentrated on the disease. The appearance and genetic relationships of the virus were unknown and the name "coronaviruses" did not yet exist.
The first coronaviruses by name were described in the mid-1960s. The first specimen discovered was the human coronavirus B814 , which was later lost . Coronaviruses are genetically highly variable ; individual species from the Coronaviridae family can also infect several species of hosts by overcoming the species barrier .
By overcoming the species barrier, infections with the SARS coronavirus ( SARS-CoV , sometimes also referred to as SARS-CoV-1) - the causative agent of the SARS pandemic 2002/2003 - as well as with the viruses that emerged in 2012, are among other things Art Middle East respiratory syndrome coronavirus ( MERS-CoV ) emerged.
By the Chinese city of Wuhan outgoing, COVID-19 pandemic is attributed to a hitherto unknown coronavirus that the name SARS-CoV-2 received.
Systematics
etymology
The name "coronaviruses" - Latin corona , wreath, crown ' - was introduced in 1968 and is related to the characteristic appearance of these viruses under the electron microscope . The first published report on the discovery gives the name given by the discoverers as based on the similarity of the enveloping fringing of the viruses to the solar corona :
“Particles are more or less rounded in profile; although there is a certain amount of polymorphism, there is also a characteristic “fringe” of projections 200 Å long, which are rounded or petal shaped, rather than sharp or pointed, as in the myxoviruses. This appearance, recalling the solar corona, is shared by mouse hepatitis virus and several viruses recently recovered from man, namely strain B814, 229E and several others. "
“The particles are more or less rounded in cross section; although there is some degree of polymorphism, there is also a distinctive “fringe” of 200 Å long appendages that are rounded or petal-shaped, rather than angular [?] or pointed like the myxoviruses. This solar corona-like appearance is shared by the mouse hepatitis virus and some recently human-derived viruses, notably B814, 229E and a few others. "
Another report traces the discoverers' choice back to the similarity of the hull edging to a crown, but refers to the first report, which gives a different description. Two virological reference works contain a chapter in which an author has the same name as a person from the above. G. Discovery group is ("Kenneth McIntosh" vs. "K. McIntosh"), and, in which the "crown etymology" is given.
structuring
The Coronaviridae family is divided into two subfamilies and currently five genera based on phylogenetic properties (a sixth is proposed). Properties such as host spectrum, organ spectrum or type of disease do not play a role in the classification.
The current two subfamilies are called Orthocoronavirinae and Letovirinae . It is Orthocoronavirinae by far the larger of the two. Letovirinae is a much younger taxon within the Coronaviridae family , and so far only one type of letovirus is known: Microhyla letovirus 1 .
The five current genera are called: Alphacoronavirus , Betacoronavirus , Deltacoronavirus , Gammacoronavirus and Alphaletovirus . Another genus “ Epsiloncoronavirus ” could be added. The former genera Torovirus and Bafinivirus are now found in the subfamily Torovirinae in the nidovirus family Tobaniviridae .
The number of species is constantly changing, especially since the SARS pandemic in 2002/2003 . Since then, research in the field of coronaviruses has increased massively. The species are classified into a large number of sub-genera. These are intended to help with the systematic classification of coronavirus types that have not yet been described in detail or will be discovered in the future and are listed in the systematic overview (see below).
Internal system
Only the most important species are given:
- Coronaviridae family
-
Subfamily Letovirinae
-
Genus Alphaletovirus
-
Milecovirus subgenus
- Species Microhyla letovirus 1 (MLeV) (*)
-
Milecovirus subgenus
-
Genus Alphaletovirus
-
Subfamily Orthocoronavirinae , formerly subfamily Coronavirinae , formerly genus Coronavirus
-
Genus Alphacoronavirus , formerly phylogroup group 1 coronaviruses
-
Colacovirus subgenus
- Species Bat coronavirus CDPHE15
-
Subgenus Decacovirus
- Species Rhinolophus ferrumequinum alphacoronavirus HuB-2013
-
Subgenus Duvinacovirus
- Species Human coronavirus 229E (German human coronavirus 229E , HCoV-229E, also occurs in bats.)
-
Subgenus Luchacovirus
- Species Lucheng Rn rat coronavirus
-
Minacovirus subgenus
- Species Ferret coronavirus
- Species Mink coronavirus 1
-
Minunacovirus subgenus
- Species Miniopterus bat coronavirus 1
- Species Miniopterus bat coronavirus HKU8 (Bat-CoV-HKU8)
-
Subgenus Myotacovirus
- Species Myotis ricketti alphacoronavirus Sax-2011
- Species Nyctalus velutinus alphacoronavirus SC-2013
-
Subgenus Pedacovirus
- Species Feline infectious peritonitis virus
- Species Porcine epidemic diarrhea virus (German Porcine Epidemic Diarrhea Virus, PEDV)
- Species Scotophilus bat coronavirus 512
-
Rhinacovirus subgenus
-
Species Rhinolophus bat coronavirus HKU2 (Bat-CoV-HKU2)
- Subspecies Swine Acute Diarrhea Syndrome Coronavirus (SADS-CoV), pathogen of SADS
-
Species Rhinolophus bat coronavirus HKU2 (Bat-CoV-HKU2)
-
Setracovirus subgenus
- Species human coronavirus NL63 (HCov-NL63, also occurs in bats.)
- Species NL63-related bat coronavirus strain BtKYNL63-9b
- Subgenus Soracovirus
- Subgenus Sunacovirus
-
Subgenus tegacovirus
-
Species Alphacoronavirus 1 (*)
- Subspecies Canine Coronavirus ( English canine coronavirus , CCoV), former species Canine coronavirus
- Subspecies Felines Coronavirus ( English feline coronavirus , FCoV, Felines Infectious Peritonitis Virus, English feline infectious peritonitis virus , FIPV), former species Feline coronavirus
- Subspecies Transmissible Gastroenteritis Virus (TGEV), former species Transmissible gastroenteritis virus
-
Species Alphacoronavirus 1 (*)
-
Colacovirus subgenus
-
Genus Betacoronavirus , formerly phylogroup group 2 coronaviruses
-
Subgenus Embecovirus
-
Species betacoronavirus 1
- Bovine Coronavirus (BCoV) subspecies
- Subspecies Equines Coronavirus (ECoV-NC99)
- Subspecies human coronavirus OC43 (HCoV-OC43, also affects chimpanzees , also occurs in rodents.)
- Subspecies Porcine Hemagglutinating Encephalomyelitis Virus (HEV)
- Subspecies Human Enteric Coronavirus (HECoV)
- Species China Rattus coronavirus HKU24 (RtCov-HKU24)
- Species human coronavirus HKU1 (HCoV-HKU1, also occurs in rodents)
-
Species Murine coronavirus (*)
- Subspecies Murine hepatitis virus ( English mouse hepatitis virus , MHV), former species Murine hepatitis virus
- Subspecies rat coronavirus ( English rat coronavirus , RtCoV), former species rat coronavirus
- Subspecies puffinosis coronavirus (PV), in black- billed shearwaters ( Puffinus puffinus ), formerly species Puffinosis coronavirus
-
Species betacoronavirus 1
-
Subgenus Hibecovirus
- Species Bat Hp-betacoronavirus Zhejiang2013
-
Subgenus Merbecovirus
- Species Hedgehog coronavirus 1
- Species Middle East respiratory syndrome-related coronavirus (German MERS-Coronavirus, MERS-CoV)
- Species Pipistrellus bat coronavirus HKU5 (Bat-CoV-HKU5)
- Species Tylonycteris bat coronavirus HKU4 (Bat-CoV-HKU4)
-
Subgenus Nobecovirus
- Species Rousettus bat coronavirus GCCDC1
- Species Rousettus bat coronavirus HKU9 (Bat-CoV-HKU9)
-
Subgenus Sarbecovirus
-
Species Severe acute respiratory syndrome-related coronavirus (dt. SARS coronavirus, English SARS-related coronavirus , SARS-CoV, SARSr-CoV), Old. Species Severe acute respiratory syndrome coronavirus (to 2009, English SARS coronavirus , dt. SARS -Coronavirus , name identical to the only subspecies at that time)
- Subspecies Severe acute respiratory syndrome coronavirus (SARS-CoV, SARS-Coronavirus, also SARS-CoV-1), pathogen of SARS
- Subspecies Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, also English 2019-novel Coronavirus , 2019-nCoV or Wuhan seafood market pneumonia virus ), pathogen of COVID-19
- Strain SARS-CoV / SARS-CoV-2 RdRp (artificial mutant: original SARS virus SARS-CoV-1 with exchanged RdRp gene from SARS-CoV-2)
- Strain Rhinolophus affinis bat coronavirus RaTG13 (BatCoV-RaTG13, Bat_SL-CoV_RaTG13), found in Java horseshoe bats( Rhinolophus affinis , English intermediate horseshoe bat ) in the Chinese province of Yunnan (with gene sequence KP876546 for BtCoV / 4991 polymerase)
- Strain Bat coronavirus RmYN01 (BatCoV-RmYN01 BetaCoV / Rm / Yunnan / YN01 / 2019)
- Strain Rhinolophus malayanus bat coronavirus RmYN02 (BatCoV-RmYN02 BetaCoV / Rm / Yunnan / YN02 / 2019), found in Malay horseshoe bat ( R. malayanus ).
- Strains of SARS-like bat coronaviruses without assignment: Bat_SL-CoV_ZC45 (bat-SL-CoVZC45), Bat_SL-CoV_ZXC21 (bat-SL-CoVZXC21), Bat_SL-CoV_Rs4231, Bat_SL-CoV_RsLsLs-Co, Bat_SL-CoV_RsLsLs-Co, Bat_SL-CoV_RsLsLs-40 CoV_Rf9402, Bat_SL-CoV_WIV16, Bat_SL-CoV_GX2013, Bat_SL-CoV_Anlong-112, Bat_SL-CoV_Shaanxi2011, Bat_SL-CoV_Yunnan2011, Bat_SL-CoV_HuB2013, Bat_SL-CoV_Longguan, Bat_SL-CoV_As6526, Bat_SL-CoV_YN2013, Bat_SL-CoV_YN2018B, Bat_SL-CoV_YN2018C, Bat_SL- CoV_HKU3-3, Bat_SL-CoV_HKU3-7, Bat_SL-CoV_HKU3-12, Bat_SL-CoV_279, Bat_SL-CoV_SC2018, Bat_SL-CoV_BM48-31, Bat_SL-CoV_BtKY72, Bat_SL-CoV_YNLF-34C, Bt SLCoV Rp3 (infected Rhinolophus sinicus ) SARS-CoV SZ3 and SZ16 (infect larval rollers).
-
suggested Species pangolin coronavirus ( Manis- CoV or English pangolin SARS-like coronavirus (Pan_SL-CoV), Pangolin-CoV)
-
Clade Pan_SL-CoV_GX found in Malaysian pangolins ( Manis javanica , English pangolin Sunda , the Chinese customs in the province G uang x i confiscated)
- Tribe Pan_SL-CoV_GX / P4L
- Trunk Pan_SL-CoV_GX / P2V
- Strain Pan_SL-CoV_GX / P1E
- Strain Pan_SL-CoV_GX / P5L
- Strain Pan_SL-CoV_GX / P5E
- Strain Pan_SL-CoV_GX / P3B
-
Klade Pan_SL-CoV_GD found in Malay pangolins ( Manis javanica , English Sunda pangolin ,confiscatedby Chinese customs in the province of G uang d ong )
- Strain Pan_SL-CoV_GD / P1La
- Trunk Pan_SL-CoV_GD / P2S
- Strain Pan_SL-CoV_GD / P1L with the SRA identifiers SRR10168374, SRR10168392, SRR10168376, SRR10168377 and SRR10168378
- Metagenome MP789 (pangolin),
-
Clade Pan_SL-CoV_GX found in Malaysian pangolins ( Manis javanica , English pangolin Sunda , the Chinese customs in the province G uang x i confiscated)
-
Species Severe acute respiratory syndrome-related coronavirus (dt. SARS coronavirus, English SARS-related coronavirus , SARS-CoV, SARSr-CoV), Old. Species Severe acute respiratory syndrome coronavirus (to 2009, English SARS coronavirus , dt. SARS -Coronavirus , name identical to the only subspecies at that time)
-
Subgenus Embecovirus
-
Genus Gammacoronavirus , formerly phylogroup group 3 coronaviruses
-
Brangacovirus subgenus
- Species Goose coronavirus CB17
-
Subgenus Cegacovirus
- Species Beluga whale coronavirus SW1 (BWCoV-SW1)
-
Igacovirus subgenus
-
Species Avian coronavirus (German bird coronavirus) (*)
- Subspecies Turkey Coronavirus (TCoV)
- Subspecies Pheasant Coronavirus (PhCoV)
- Subspecies Infectious Bronchitis Virus ( English avian infectious bronchitis virus , IBV), causative agents of infectious bronchitis in birds
-
Species Avian coronavirus (German bird coronavirus) (*)
-
Brangacovirus subgenus
-
Genus Deltacoronavirus
-
Subgenus Andecovirus
- Species Wigeon coronavirus HKU20 (WiCoV-HKU20)
-
Subgenus Buldecovirus (including former subgenus Moordecovirus )
- Species Bulbul coronavirus HKU11 (BuCoV-HKU11) (*)
- Species coronavirus HKU15
- Species Munia coronavirus HKU13 (German bronze male coronavirus HKU13, MunCoV HKU13)
- Species Common moorhen coronavirus HKU21 (CMCoV_HKU21, previously in the former subgenus Moordecovirus )
- Species White-eye coronavirus HKU16
- Species Thrush coronavirus HKU12 (German thrush coronavirus HKU12, ThCoV-HKU12)
- suggested Species Sparrow coronavirus HKU17 (German sparrow coronavirus HKU17, SpCoV-HKU17)
-
Herdecovirus subgenus
- Species Night heron coronavirus HKU19
-
Subgenus Andecovirus
-
suggested Genus " epsilon coronavirus "
-
suggested Subgenus " Tropepcovirus "
- suggested Species " Tropidophorus coronavirus 118981 " (Tsin-CoV 118981, also " Guangdong chinese water skink coronavirus "), in Chinese water skink ( Tropidophorus sinicus ) (*)
-
suggested Subgenus " Tropepcovirus "
-
Genus Alphacoronavirus , formerly phylogroup group 1 coronaviruses
Subfamily Torovirinae (including genera Torovirus and Bafinivirus )→ Family Tobaniviridae
In the following cladogram according to Mang Shi et al. (2016) the names were updated according to ICTV MSL # 35 (as of March 2020):
| Coronaviridae |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
External system
order Subordination family Nidovirals Abnidovirineae Abyssoviridae Arnidovirineae Arteriviridae Cremegaviridae Gresnaviridae Olifoviridae Cornidovirineae Coronaviridae Mesnidovirineae Medioniviridae Mesoniviridae Monidovirineae Mononiviridae Nanidovirineae Nanghoshaviridae Nanhypoviridae Ronidovirineae Euroniviridae Roniviridae Tornidovirineae Tobaniviridae
Taxonomic backgrounds
Subfamilies
Until 2018, Coronaviridae consisted of the subfamilies Coronavirinae and Torovirinae . Before that it was bigeneric, consisting of the genera Coronavirus and Torovirus .
In 2009, in the course of the further development of the family, the genus Coronavirus was raised to the subfamily Coronavirinae . The subfamily contained the same viruses as the genus before. It had been placed next to the new subfamily Torovirinae , which included the genus Torovirus .
In 2018 the subfamily Coronavirinae was renamed Orthocoronavirinae . More in the section Orthocoronavirinae .
Genera
The viruses within the old genus Coronavirus had been divided into three non-taxonomic, monophyletic groups ( clades ) based on phylogenetic and serological properties of the species , which were formerly known as HCoV-229E-like (group 1), HCoV-OC43 -like (group 2) and IBV-like (group 3) coronaviruses. Group 2 was further divided into the four likewise monophyletic subgroups 2A to 2D.
During the formation of the new subfamily Coronavirinae from the old genus Coronavirus in 2009 , the three informal, but long and well-established monophyletic groups became today's genera Alpha- to Gammacoronavirus (in the order of the Greek alphabet: Alpha, Beta, Gamma, Delta , Epsilon, ...). The genus Deltacoronavirus was added later, another genus " Epsiloncoronavirus " could soon be added. The genus Torovirus , however, remained under this name and was classified in the new subfamily Torovirinae . In addition to torovirus , the new genus Bafinivirus of bacilliform (rod-shaped) fish viruses was added to this.
In 2018, the toroviruses disappeared completely from the Coronaviridae family , and the letoviruses were added at the same time.
With "Toroviruses" are meant here all viruses of the subfamily Torovirinae in the form at that time. This subfamily included the viruses of the genera Toro and Bafinivirus and other viruses. In 2018, these viruses were placed in the new nidovirus family Tobaniviridae with a newly designed subfamily structure, which ended ambiguities among other things: Now only the viruses of the genus Torovirus are also classified as Toroviruses .
| Genera / generic groups | Family affiliation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| until 2009 | from 2009 on | from 2011 | from 2018 | coming (as of July 2020) | until 2018 | from 2018 | ||||
| Coronavirus |
|
|
Alphacoronavirus | Coronaviridae | Coronaviridae | |||||
|
|
Beta coronavirus | ||||||||
|
|
Gamma coronavirus | ||||||||
| Delta Coronavirus | ||||||||||
| " Epsilon coronavirus " | (n / A.) | |||||||||
| Alphaletovirus | ||||||||||
| Torovirus | Torovirus | Coronaviridae | Tobaniviridae | |||||||
| Bafinivirus | ||||||||||
Legend: monophyletic groups used similarly to the genus, subfamilies
Subgenera
In 2018, for the first time, a number of subgenera were defined in the Coronaviridae family . Including sub-genera in the genus Betacoronavirus (see ibid.). Just as this genus had previously been formed from the well-known phylogroup 2, the subgroups 2A to 2D now known under the names line A to line D were also converted into sub-genres.
In 2018, for the first time, a number of subgenera were defined in the Coronaviridae family . The name of the sub-genera corresponds to a scheme of descriptive names in the form of neologisms . They are all suitcase words (according to the ICTV code a so-called siglum ( English / Latin )) such as B. Sarbecovirus corresponding to SAR S-like be ta co rona virus .
An express reason for this was a classification backlog caused by many viruses that have not yet been described in detail and therefore still have to be classified. Many new discoveries and new descriptions were to be expected as a result of the great advances in genomany analysis and the increased research activity in the field of coronaviruses since the SARS pandemic of 2002/2003 . With this aspect of the taxonomic structure, the aim was to offer a rational framework for the systematic classification that only focuses on the genomic properties of the viruses. Because they had long been determined as the only relevant aspect for the classification.
The sub-genera also include those in the genus Betacoronavirus (see ibid.). Just as this genus had previously been formed from the well-known phylogroup 2, the subgroups 2A to 2D now known under the names line A to line D were also converted into sub-genres.
meaning
medicine
Diseases
Coronaviruses cause very different diseases in different vertebrates such as mammals , birds , fish and frogs .
In humans, various types of coronavirus are important as pathogens ranging from light respiratory infections (colds) to severe acute respiratory syndrome . A causal involvement in gastroenteritis is possible, but clinically and numerically it does not play a major role. A total of seven human pathogenic coronaviruses are known (as of February 2020): in addition to SARS-CoV (-1), SARS-CoV-2 and MERS-CoV, HCoV-HKU1 , HCoV-OC43 (all belonging to the genus Betacoronavirus ), HCoV-NL63 and HCoV -229E (both belonging to the genus Alphacoronavirus ). The last four are responsible for about 5-30% of all acute respiratory diseases and typically lead to rhinitis , conjunctivitis , pharyngitis , occasionally a laryngitis or otitis media (otitis media). Infections of the lower respiratory tract are also possible, especially in the case of co-infections with other respiratory pathogens (such as rhinoviruses , enteroviruses , respiratory syncytial viruses (RSV), parainfluenza viruses ). Serious disease courses are observed above all in pre-existing diseases, especially of the cardiopulmonary system, and in connection with transplants ( immunosuppression ); usually only comparatively minor symptoms occur.
In a follow-up study over eight years - before the outbreak of COVID-19 - in selected households with around a thousand people in Michigan , USA, almost 1000 acute respiratory diseases were caused by HCoV infections (mostly OC43). The seasonally limited occurrence of these infections in the months of December to April / May was noticeable, but this cannot necessarily be assumed for SARS-CoV-2.
Subfamilies
The subfamily was created in 2018 from the renaming of the subfamily Coronavirinae . This in turn arose when the genus Coronavirus was raised to a subfamily in 2009 (and the ending -virus was changed to -virinae ).
In parallel, until 2018 there was a torovirus sister taxon with an analogous name within the Coronaviridae family : Torovirus as a sister genus of Coronavirus and Torovirinae as a sister subfamily of Coronavirinae . This resulted in the situation that toroviruses were both coronaviruses by name, because they belonged to the Coronaviridae family , and non-coronaviruses, because they did not belong to the subfamily / subgenus Coronavirinae / Coronavirus . Such a thing was never unusual in virus taxonomy. Similar entanglements existed at times between toroviruses and bafiniviruses.
The toroviruses did not correspond to the name "coronavirus" or only to a limited extent.
The idea behind the name “Coronavirus ” is a sun-like basic shape surrounded by a sun-like corona . This refers to a virus that appears to be disk-shaped in electron microscopic images and is framed by a clear, separate ring of club-shaped or petal-shaped projections. (See also section Etymology ) But neither the Toro nor the Bafiniviruses were disc or spherical. (Although toroviruses are said to have been made to produce spherical virions in cell culture.) They got their name precisely because they were torus or bacillus-shaped, i.e. ring-shaped or rod-shaped. The toroviruses were actually rod-shaped structures that were bent into an almost closed ring or a crescent-shaped shape.
In addition to the differences, there were also deviations in the genome structure between toro and bafiniviruses on the one hand and coronaviruses on the other. The genome encoded e.g. B. not the envelope protein E of the other coronaviruses and was organized much more simply overall. In particular, almost all of the typical auxiliary genes for the other coronaviruses were missing.
Toro and Bafiniviruses also had tubular, helical nucleocapsids , while those of the other coronaviruses were loosely wound. The capsid proteins were also less than half the size of those of the other coronaviruses. Ultimately, the Bafiniviruses also differed in the host spectrum (= aquatic) from the viruses within Coronavirinae / Coronavirus (= terrestrial).

Due to these differences, the viruses of the then subfamily Coronavirinae or the former genus Coronavirus were typically referred to as "real coronaviruses" until 2018 . This is how they were distinguished from the merely formal corona viruses from the group of toro and bafiniviruses. Then in 2018 the toro and bafiniviruses from the Coronaviridae family were removed . At the same time, the name was changed from Coronavirinae to today's subfamily Orthocoronavirinae . Since then, the well-defined name "orthocoronaviruses" has been available for the viruses in this group.
From a formal point of view, the name "coronaviruses" now designates all viruses of the Coronaviridae family and - if they are single - nothing else. This also corresponds to the ICTV rules of virus taxonomy, which forbids ambiguities in the (re) naming of taxa.
- So-called. "Real" coronaviruses
- Corona viruses also until 2018
Type of torovirus (here BEV , type Equine torovirus )
Type of Bafinivirus (here WBV , type White bream virus )
Since the Letoviruses that were added as part of the name change are much more similar to the orthocoronaviruses, the name "Coronaviruses" is now also applicable to all viruses of the Coronaviridae family and there would no longer have to be any ambiguity. Nevertheless, the viruses of the subfamily Orthocoronavirinae are still primarily referred to as the coronaviruses. This may be due to the still little research on the Letovirus group (subfamily Letovirinae ).
Letovirinae
The subfamily was created in 2018 due to the discovery of the frog virus species Microhyla letovirus 1 .
Not much is known about the Letoviruses as a group, as they have so far only been represented by this one species that has not yet been very thoroughly researched. They agree with the orthocoronaviruses in all essential properties, but in statistical relationship analyzes of the genomes they form an independent group that is too far removed from the orthocoronaviruses to form a new genus within the subfamily Orthocoronavirinae alongside the other genera present there.
A possible close relative of Microhyla letovirus 1 is the proposed and so far unclassified nidovirus species " Pacific salmon nidovirus " (PsNV). It is likely that within the Coronaviridae family it will belong to a genus of its own parallel to Alphaletovirus . Whether within the same subfamily Letovirinae (another Letovirus), or beyond it is still unclear.
literature
- David M. Knipe, Peter M. Howley (eds.-in-chief): Fields' Virology. 5th edition, 2 volumes, Lippincott Williams & Wilkins, Philadelphia 2007, ISBN 0-7817-6060-7 .
- CM Fauquet, MA Mayo, et al. : Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London / San Diego 2005, ISBN 0-12-249951-4 .
- AMQ King, MJ Adams, EB Carstens, EJ Lefkowitz (Eds.): Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier, Amsterdam et al. 2012, ISBN 978-0-12-384684-6 , pp. 806-828.
- S. Modrow, D. Falke, U. Truyen: Molecular Virology . Spectrum textbook, 2nd edition, Akademischer Verlag, Heidelberg / Berlin 2003, ISBN 3-8274-1086-X , pp. 214–226.
- PS Masters: The molecular biology of coronaviruses. In: Advances in virus research. Volume 66, 2006, pp. 193-292, doi: 10.1016 / S0065-3527 (06) 66005-3 (free full text access ), PMID 16877062 .
See also
- June Almeida took the first pictures of coronaviruses with an electron microscope
Web links
- How dangerous is the novel corona virus? - Hamburger Abendblatt from February 5, 2020.
- Alex Knapp: The secret history of the first coronavirus 229E , in Forbes from April 12, 2020 - Research history of human pathogenic coronaviruses (229E, OC43, HKU1, NL63, IBV)
- Jürgen Schönstein: Reading tip: Drosten portrait in science , on: scienceblogs.de - ditto: Zeitschiene
Individual evidence
- ↑ ICTV Master Species List 2018b v2 . MSL # 34v, March 2019.
- ↑ a b c d e f g h ICTV: ICTV Taxonomy history: Severe acute respiratory syndrome-related coronavirus. EC 51, Berlin, Germany, July 2019; Email ratification March 2020 (MSL # 35)
- ^ Jens H. Kuhn, Peter B. Jahrling: Clarification and guidance on the proper usage of virus and virus species names . In: Archives of Virology . tape 155 , April, p. 445–453, March 4, 2010, Table 1, pp. 12 , Z e 1, S p 3 , doi : 10.1007 / s00705-010-0600-9 , PMID 20204430 , PMC 2878132 (free full text) - (English, full text [PDF; 64 kB ; accessed on June 18, 2020]): “Taxa refer to theoretical averages of groups of viruses; hence taxa are not preceded by articles. "
- ↑ Fact check on coronavirus myths - Of chlorine dioxide, cover-up and onions . on: n-tv.de from March 18, 2020. Source: RKI
- ↑ Denise Gellene: Overlooked No More: June Almeida, Scientist Who Identified the First Coronavirus . On: nytimes.com from May 8, 2020 (update May 10, 2020); accessed on August 5, 2020.
- ↑ B. Delmas, H. Laude: Assembly of Coronavirus Spike Protein Into Trimers and Its Role in Epitope Expression. In: Journal of Virology. Volume 64, No. 11, November 1990, PMID 2170676 , PMC 248586 (free full text), pp. 5367-5375.
- ^ Structure of "Spike" Protein in New Coronavirus Determined. On: ChemistryViews.org February 21, 2020; accessed on March 3, 2020.
- ↑ Spike Protein / S Protein . on Sino Biological.com ; accessed on March 3, 2020.
- ^ A b M. R. Denison, R. L. Graham, E. F. Donaldson, L. D. Eckerle, R. S. Baric: Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. In: RNA Biology. Volume 8, No. 2, March-April 2011, pp. 270-279, doi: 10.4161 / rna.8.2.15013 , ISSN 1555-8584 , PMID 21593585 , PMC 3127101 (free full text).
- ↑ P. C. Y. Woo, M. Wang, S. K. P. Lau, H. Xu, R. W. S. Poon, R. Guo, B. H. L. Wong, K. Gao, H.-w. Tsoi, Y. Huang, K.S.M. Li, C.S.F. Lam, K.-h. Chan, B.-j. Zheng, K.-y. Yuen: Comparative Analysis of Twelve Genomes of Three Novel Group 2c and Group 2d Coronaviruses Reveals Unique Group and Subgroup Features . In: Journal of Virology . tape 81 , no. 4 , February 2007, p. 1574–1585 , doi : 10.1128 / JVI.02182-06 , PMID 17121802 , PMC 1797546 (free full text).
- ↑ a b c d e David Cyranoski: Virology: Portrait of a killer . Online edition of the article in Spektrum der Wissenschaft. No. 8, August 2020, pp. 40–49.
- ↑ C. B. Hudson, F. R. Beaudette: INFECTION OF THE CLOACA WITH THE VIRUS OF INFECTIOUS BRONCHITIS . In: Science . tape 76 , no. 1958 . American Association for the Advancement of Science, July 8, 1932, ISSN 0036-8075 , pp. 34 , doi : 10.1126 / science.76.1958.34-a , PMID 17732084 (English, full text [accessed on August 10, 2020]).
- ↑ Yassine Kasmi, Khadija Khataby, Amal Souiri, Moulay Mustapha Ennaji: Coronaviridae: 100,000 Years of Emergence and Reemergence . Book chapter. In: Moulay Mustapha Ennaji (Ed.): Emerging and Reemerging Viral Pathogens . tape 1: Fundamental and Basic Virology Aspects of Human, Animal and Plant Pathogens . Elsevier / Academic Press, 2020, ISBN 978-0-12-819400-3 , pp. 127–149 , doi : 10.1016 / B978-0-12-819400-3.00007-7 (English, full text [PDF; 2.6 MB ; accessed on August 10, 2020]).
- ↑ Fact check on coronavirus myths - Of chlorine dioxide, cover-up and onions . on: n-tv.de from March 18, 2020. Source: RKI
-
↑
Jeffrey Kahn, Kenneth McIntosh: History and Recent Advances in Coronavirus Discovery. In: The Pediatric Infectious Disease Journal. Volume 24, No. 11, 2005, pp. S223-S227, doi: 10.1097 / 01.inf.0000188166.17324.60 .
National Foundation for Infectious Diseases: Coronaviruses. On: nfid.org ; accessed on March 23, 2020. -
↑ New-type coronavirus causes pneumonia in Wuhan: expert. On: xinhuanet.com January 9, 2020.
WHO: Pneumonia of unknown cause - China. Disease Outbreak News from January 5, 2020. - ↑ Hamburger Abendblatt- Hamburg: How dangerous is the new type of corona virus? February 5, 2020, accessed on March 18, 2020 (German).
- ↑ Virology: Coronaviruses . In: Nature . tape 220 , November 16, 1968, ISSN 1476-4687 , p. 650 , doi : 10.1038 / 220650b0 (English, full text [PDF; 1.8 MB ; accessed on June 18, 2020] First report on the discovery of the coronavirus).
- ^ A. F. Bradburne: Antigenic Relationships amongst Coronaviruses . In: Archives for Virus Research . tape 31 . Springer-Verlag, February 6, 1970, p. 352–364 , doi : 10.1007 / BF01253769 (English, full text [PDF; 923 kB ; accessed on June 18, 2020]).
- ↑ Kenneth McIntosh (for this chapter): Feigin and Cherry's Textbook of Pediatric Infectious Disease . 6th edition. Saunders / Elsevier, Philadelphia, PA (USA) 2009, ISBN 978-1-4160-4044-6 , section XVII, subsection 10, chapter 189A. Coronaviruses and Toroviruses , S. 2380 (English, full text of the chapter [PDF; 265 kB ; accessed on July 7, 2020]): "[T] he genus Coronavirus of the family Coronaviridae , named for the crown-like appearance of their surface projections on electron microscopy."
- ↑ Kenneth McIntosh, Stanley Perlman (both for this chapter): Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases . 8th edition. tape 2 . Saunders / Elsevier, Philadelphia, PA (USA) 2014, ISBN 978-1-4557-4801-3 , chapter 157. Coronaviruses, Including Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) , p. 1928 , right column (English, 3904 p., Full text of the chapter [PDF; 1.5 MB ; accessed on July 8, 2020]): "[T] he name coronavirus (the prefix corona denoting the crownlike appearance of the surface projections) was chosen to signify this new genus."
-
↑ a b c
- Raoul J. de Groot, John Ziebuhr, Leo L. Poon, Patrick C. Woo, Pierre Talbot, Peter JM Rottier, Kathryn V. Holmes, Ralph Baric, Stanley Perlman, Luis Enjuanes, Alexander E. Gorbalenya: Revision of the family Coronaviridae . Proposal. In: Virus Taxonomy . History. Revision 2009, June. International Committee on Taxonomy of Viruses (ICTV), 2008, Proposal Code 2008.085-126V (English, ictvonline.org [PDF; 175 kB ; accessed on May 5, 2020]): “The naming of coronavirus genera is according to the Greek alphabet. The viruses grouped in currently recognized genera form distinct monophylogenetic clusters, but do not share other obvious traits (host tropism, organ tropism, type of disease) to suggest a common denominator. Hence, the CSG proposes to use a neutral nomenclature, [...]. "
- Authors of the ninth ICTV report: Virus Taxonomy - Classification and Nomenclature of Viruses. Online edition. Cape. " Coronaviridae ". In: ICTV Reports . International Committee on Taxonomy of Viruses, 2011, accessed on June 12, 2020 (English, archived in parallel on April 2, 2019 on web.archive.org .): “Viruses that share more than 90% aa sequence identity in the conserved replicase domains are considered to belong to the same species. This 90% identity threshold serves as the sole species demarcation criterion. "
- Proposal for ICTV revision № 2018a: 2017.013S. (PDF in ZIP folder; 4.49 MB) In: ICTV homepage . International Committee on Taxonomy of Viruses (ICTV), accessed on May 7, 2020 (English): "We would like to stress that we expect the proposed taxonomy structure to provide a framework for the rationalization of the molecular and biological properties of viruses in these two families, which, in many cases, remain to be determined and, therefore, cannot be used to evaluate the validity of the proposed structure. "
- ↑ a b c d e f g h i j k J. Ziebuhr, RS Baric, S. Baker, RJ de Groot, C. Drosten, A. Gulyaeva, BL Haagmans, BW Neuman, S. Perlman, LLM Poon, I. Sola, AE Gorbalenya: 2017.012_015S.A.v1.Nidovirales . In: Virus Taxonomy . History. Revision 2018a, July. International Committee on Taxonomy of Viruses (ICTV), February 18, 2017, Proposal Code 2017.012_015S ( ictvonline.org [ ZIP ; 5.1 MB ; accessed on May 7, 2020]).
- ↑ a b c d e f J. Ziebuhr, S. Baker, RS Baric, RJ de Groot, C. Drosten, A. Gulyaeva, BL Haagmans, BW Neuman, S. Perlman, LLM Poon, I. Sola, AE Gorbalenya: 2019.021S . Proposal document in ZIP. In: International Committee on Taxonomy of Viruses (Ed.): Virus Taxonomy . History. Revision 2019, document name in ZIP: 2019.021SAv1.Corona_Tobaniviridae.docx . EC 51, Berlin April 2019, Proposal Code 2019.021S ( ictvonline.org [ ZIP ; 1.8 MB ; accessed on June 9, 2020] ratified in March 2020, not yet fully published, proposal website on ictvonline.org ).
- ↑ a b c Khulud Bukhari, Geraldine Mulley, Anastasia A. Gulyaeva, Lanying Zhao, Guocheng Shu, Jianping Jiang, Benjamin W. Neuman: Description and initial characterization of metatranscriptomic nidovirus-like genomes from the proposed new family Abyssoviridae, and from a sister group to the _Coronavirinae_, the proposed genus Alphaletovirus . In: Virology . tape 524 . Elsevier Inc., November 2018, p. 160–171 , doi : 10.1016 / j.virol.2018.08.010 , PMID 30199753 , PMC 7112036 (free full text) - (English, full text [PDF; 3.3 MB ; accessed on May 18, 2020] "Coronavirinae": today "Orthocoronavirinae" ).
- ↑ Proposal for ICTV revision № 2018a: 2017.013S. (PDF in ZIP folder; 4.49 MB) In: ICTV homepage . International Committee on Taxonomy of Viruses (ICTV), accessed May 7, 2020 .
- ↑ a b c d e f g h i j k l ICTV : Revision of the family Coronaviridae . Taxonomic proposal to the ICTV Executive Committee 2008.085-126V.
- ^ New coronavirus emerges from bats in China, devastates young swine . On: ScienceDaily. from April 4, 2018.
- ↑ Zhou et al : Fatal swine acute diarrhea syndrome caused by an HKU2-related coronavirus of bat origin . In: Nature research, life sciences reporting summary, Letter. June 2017, doi: 10.1038 / s41586-018-0010-9 .
- ↑ ICTV: ICTV Taxonomy history: NL63-related bat coronavirus strain BtKYNL63-9b . EC 51, Berlin, Germany, July 2019; Email ratification March 2020 (MSL # 35)
- ↑ Turmeric Could have antiviral properties . On: eurekalert.org of July 17, 2020, source: Microbiology Society.
- ↑ Nadja Podbregar: Coronavirus: Are great apes also endangered? . On: scinexx.de from March 30, 2020.
- ^ NCBI: Council coronavirus . (no rank)
- ↑ Ben Hu, Lei-Ping Zeng, Xing-Lou Yang, Xing-Yi Ge, Wei Zhang, Bei Li, Jia-Zheng Xie, Xu-Rui Shen, Yun-Zhi Zhang, Ning Wang, Dong-Sheng Luo, Xiao- Shuang Zheng, Mei-Niang Wang, Peter Daszak, Lin-Fa Wang, Jie Cui, Zheng-Li Shi; Christian Drosten (Ed.): Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus . In: PLOS Pathogens. dated November 30, 2017, doi: 10.1371 / journal.ppat.1006698 .
- ↑ a b Kristian G. Andersen, Andrew Rambaut, W. Ian Lipkin, Edward C. Holmes, Robert F. Garry: The Proximal Origin of SARS-CoV-2 . On: virologica.org , source: ARTIC Network, February 17, 2020.
- ↑ a b Chengxin Zhang, Wei Zheng, Xiaoqiang Huang, Eric W. Bell, Xiaogen Zhou, and Yang Zhang: Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1 . In: Journal of Proteome Research . tape 19 , no. 4 . American Chemical Society, March 22, 2020, p. 1351–1360 , doi : 10.1021 / acs.jproteome.0c00129 (English, full text [PDF; 7.1 MB ; retrieved on June 6, 2020] In semi-italics: Manis- CoV, apparently with regard to host animal typography: Manis javanica ): “2019-nCoV and the Manis coronavirus ( Manis- CoV)”
- ↑ a b Tommy Tsan-Yuk Lam, Marcus Ho-Hin Shum, Hua-Chen Zhu, Yi-Gang Tong, Xue-Bing Ni, Yun-Shi Liao, Wei Wei, William Yiu-Man Cheung, Wen-Juan Li, Lian -Feng Li, Gabriel M. Leung, Edward C. Holmes, Yan-Ling Hu, Yi Guan: Identification of 2019-nCoV related coronaviruses 1 in Malayan pangolins in southern China . on: bioRxiv from February 18, 2020, doi: 10.1101 / 2020.02.13.945485 (preprint), doi: 10.1038 / s41586-020-2169-0 (newer version: "Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins")
- ↑ Andrea J. Pruijssers et al. : Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice . In: Cell Reports. from 7 July 2020, doi: 10.1016 / j.celrep.2020.107940 (free pre-proof)
- ↑ Tina Hesman Saey: Remdesivir may work even better against COVID-19 than we thought . On: ScienceNews of July 13, 2020
- ↑ a b c Alexandre Hassanin: Coronavirus Could Be a 'Chimera' of Two Different Viruses, Genome Analysis Suggests . On: science alert of March 24, 2020 (source: THE CONVERSATION).
- ↑ Kristian G. Andersen, Andrew Rambaut, W. Ian Lipkin, Edward C. Holmes, Robert F. Garry: The Proximal Origin of SARS-CoV-2. In: virological.org , Source: ARTIC Network, February 17, 2020, Nature
- ↑ NCBI: Bat coronavirus RaTG13 .
- ↑ a b c d Xiaojun Li, Elena E. Giorgi, Manukumar Honnayakanahalli Marichannegowda, Brian Foley, Chuan Xiao, Xiang-Peng Kong, Yue Chen, S. Gnanakaran, Bette Korber, Feng Gao: Emergence of SARS-CoV-2 through recombination and strong purifying selection . In: Science Advances, AAAs. from May 29, 2020, eabb9153, doi: 10.1126 / sciadv.abb9153
- ↑ a b c d Jose Halloy, Erwan Sallard, José Halloy, Didier Casane, Etienne Decroly, Jacques van Helden: Tracing the origins of SARS-COV-2 in coronavirus phylogenies , in: HAL from July 16, 2020, HAL Id: hal -02891455 (preprint)
- ↑ a b c Hong Zhou, Xing Chen, Tao Hu, Juan Li, Hao Song, Yanran Liu, Peihan Wang, Di Liu, Jing Yang, Edward C. Holmes, Alice C. Hughes, Yuhai Bi, Weifeng Shi: A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1 / S2 cleavage site of the spike protein . In: Current Biology. May 11, 2020 (pre-proof), doi: 10.1016 / j.cub.2020.05.023 .
- ↑ Jacinta Bowler: Researchers Find Another Virus in Bats That's Closely Related to SARS-CoV-2 . On: science alert from May 12, 2020
- ↑ Dezhong Xu, Huimin Sun, Haixia Su, Lei Zhang; Jingxia Zhang, Bo Wang, Rui Xu: SARS coronavirus without reservoir originated from an unnatural evolution, experienced the reverse evolution, and finally disappeared in the world . In: Chinese Medical Journal. Volume 127, No. 13, July 5, 2014, pp. 2537-2542, doi: 10.3760 / cma.j.issn.0366-6999.20131328
- ↑ Ming Wang et al. : SARS-CoV Infection in a Restaurant from Palm Civet . In: Emerging Infectious Diseases journal. (Emerg Infect Dis.) Volume 11, No. 12, December 2005, pp. 1860-1865, doi: 10.3201 / eid1112.041293 , PMC PMC3367621 (free full text), PMID 16485471
- ↑ NCBI: Pangolin coronavirus . (species)
- ↑ Samantha Black: Scientists use bioinformatics to investigate origin of SARS-CoV-2. (PDF) In: The Science Advisory Board . March 27, 2020, accessed on June 6, 2020 (English, in semi-italics: Manis- CoV, apparently with regard to host animal typography: Manis javanica ): “[…] the researchers drafted a genome for Manis- CoV that was used in comparison to SARS-CoV-2 using metagenomic samples. "
- ↑ Xiaojun Li, Elena E. Giorgi, Manukumar Honnayakanahalli Marichannegowda, Brian Foley, Chuan Xiao, Xiang-Peng Kong, Yue Chen, S. Gnanakaran, Bette Korber, Feng Gao: Emergence of SARS-CoV-2 through recombination and strong purifying selection . In: Science Advances - AAAs . Article no. fig9153. American Association for the Advancement of Science, May 29, 2020, ISSN 2375-2548 , doi : 10.1126 / sciadv.abb9153 (English, full text [PDF; 3.1 MB ; accessed on June 7, 2020]): “These pangolin SARS-like CoVs (Pan_SL-CoV) form two distinct clades corresponding to their locations of origin: the first clade, Pan_SL-CoV_GD, sampled from Guangdong (GD) province in China, […] The second clade, Pan_SL-CoV_GX, sampled from Guangxi (GX) province […]. ”
- ↑ NCBI Sequence Read Archive (SRA).
- ↑ NCBI: Virome of dead pangolin individuals: SRR10168376 .
- ↑ a b c Chengxin Zhang et al. : Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1 . In: American Chemical Society: J. Proteome Res. Of March 22, 2020, doi: 10.1021 / acs.jproteome.0c00129 ; PrePrint , PrePrint full text (PDF) from February 8, 2020.
- ↑ NCBI: Viromes of dead pangolin individuals: SRR10168377 .
- ↑ a b Xingguang Li, Junjie Zai, Qiang Zhao, Qing Nie, Yi Li, Brian T. Foley, Antoine Chaillon: Evolutionary history, potential intermediate animal host, and cross ‐ species analyzes of SARS ‐ CoV ‐ 2 . In: Journal of Medical Virology. Volume February 27, 2020, doi: 10.1002 / jmv.25731 , PDF , PMID 32104911 , researchgate
- ↑ NCBI: Viromes of dead pangolin individuals: SRR10168378 .
-
↑ a b
database entry and relevant proposal:
- Entry: ICTV Taxonomy history: Moordecovirus . In: ICTV homepage . International Committee on Taxonomy of Viruses (ICTV), accessed June 9, 2020 .
- Proposal 2019.021S to ICTV -Revision № 2019: Create eight new species in the subfamily Orthocoronavirinae of the family Coronaviridae and four new species and a new genus in the subfamily Serpentovirinae of the family Tobaniviridae . (One of several PDFs in ZIP folder; 725 kB) 2019.021S. In: ICTV homepage . International Committee on Taxonomy of Viruses (ICTV), accessed June 9, 2020 .
- ↑ Spreadsheet 2017.012-015S on ICTV revision № 2018a: 2017.012-015S. (An XLSX file in ZIP folder; 4.49 MB) In: ICTV homepage . International Committee on Taxonomy of Viruses (ICTV), accessed June 9, 2020 .
- ↑ Sander van Boheemen: Virus Discovery and Characterization using next-generation sequencing . Proefschrift Erasmus Universiteit Rotterdam, Rotterdam 2014, ISBN 978-90-8891-932-9 , Figure 3.
- ↑ NCBI: Thrush coronavirus HKU12 . (species)
- ↑ NCBI: Sparrow coronavirus HKU17 . (species)
- ↑ a b Mang Shi, Xiabn-Dan Lin, Jun-Huia Tian et al. : Redefining the invertebrate RNA virosphere . In: Nature 540, pp. 539-543, November 23, 2016, doi: 10.1038 / nature20167 , via ResearchGate (full text), Supplement (PDF) .
- ↑ a b Strain Details for Guangdong chinese water skink coronavirus Strain ZGLXR118981. In: ViPR homepage . May 2, 2020, accessed June 16, 2020 .
- ↑ NCBI: Guangdong chinese water skink coronavirus (species)
- ↑ a b c d e f g Raoul J. de Groot, John Ziebuhr, Leo L. Poon, Patrick C. Woo, Pierre Talbot, Peter JM Rottier, Kathryn V. Holmes, Ralph Baric, Stanley Perlman, Luis Enjuanes, Alexander E. Gorbalenya: Revision of the family Coronaviridae . Proposal. In: Virus Taxonomy . History. Revision 2009, June. International Committee on Taxonomy of Viruses (ICTV), 2008, Proposal Code 2008.085-126V (English, ictvonline.org [PDF; 175 kB ; accessed on May 5, 2020]).
- ↑ a b c d e f g h Authors of the ninth ICTV report: Virus Taxonomy - Classification and Nomenclature of Viruses. Online edition. Cape. " Coronaviridae ". In: ICTV Reports . International Committee on Taxonomy of Viruses, 2011, accessed on June 12, 2020 (English, parallel archived on April 2, 2019 on web.archive.org .).
- ↑ Kenneth McIntosh (for this chapter): Feigin and Cherry's Textbook of Pediatric Infectious Disease . 6th edition. Saunders / Elsevier, Philadelphia, PA (USA) 2009, ISBN 978-1-4160-4044-6 , section XVII, subsection 10, chapter 189A. Coronaviruses and Toroviruses , S. 2380 (English, full text of the chapter [PDF; 265 kB ; accessed on July 7, 2020] In addition to the quotation, one or more uses in the chapter (including references) of: "HCoV-229E-like isolates", "HCoV-OC43-like isolates", "HCoV-229E-like viruses", "HCoV-229E-like strains", "HCoV-OC43-like strains", "229E-like coronavirus", "" avian infectious bronchitis virus-like "viruses (coronaviruses)", "" IBV-like "virus", " “IBV-like” viruses ”.): “ When sufficient virus has been recovered for characterization, most isolates have proved to be similar, either HCoV-229E-like or HCoV-OC43-like, 57 although several HCoVs, including the very first isolate HCoV-B814, remain antigenically uncharacterized. [... next column ...] The antigenic interrelationships of these four proteins have permitted arrangement of both the animal coronaviruses and HCoVs into three groups. The two known HCoV serotypes, each along with several other mammalian coronaviruses, have been placed in group I (HCoV-229E) or II (HCoV-OC43), and avian infectious bronchitis virus is the single member of group III. "
- ↑ Francesca Rovida, Elena Percivalle, Maurizio Zavattoni, Maria Torsellini, Antonella Sarasini, Giulia Campanini, Stefania Paolucci, Fausto Baldanti, M. Grazia Revello, Giuseppe Gerna: Monoclonal antibodies versus reverse transcription ‐ PCR for detection of respiratory viruses in a patient population with respiratory tract infections admitted to hospital . In: Journal of Medical Virology . tape 75 , edition 2, February 2005, p. 336-347. Wiley-Liss, December 15, 2004, p. 336 , left column , doi : 10.1002 / jmv.20276 , PMID 15602736 , PMC 7166428 (free full text) - (English, full text [PDF; 278 kB ; accessed on July 8, 2020]): “[H] uman coronavirus (hCoV) groups I (229E ‐ like) and II (OC43 ‐ like) […] were searched for by RT ‐ PCR […].”
-
↑
Database entry and associated revision proposal:
- E INTRAG : ICTV taxonomy history: Delta coronavirus . In: Virus Taxonomy . International Committee on Taxonomy of Viruses , accessed May 22, 2020 .
- P ROPOSAL : Raoul J. de Groot and Alexander E. Gorbalenya: A new genus and three new species in the subfamily Coronavirinae . Proposal. In: International Committee on Taxonomy of Viruses (Ed.): Virus Taxonomy . History. Revision 2011. EC43, Sapporo September 2011, Proposal Code 2010.023a-dV (English, ictvonline.org [PDF; 220 kB ; accessed on June 17, 2020] First edition: EC 42, Paris 2010).
- ↑ a b c ICTV Code - The International Code of Virus Classification and Nomenclature. In: ICTV homepage . International Committee on Taxonomy of Viruses, October 2018, accessed June 16, 2020 .
- ↑ a b Prof. Dr. John Ziebuhr: Medical microbiology and infectious diseases . Ed .: Sebastian Suerbaum, Gerd-Dieter Burchard, Stefan HE Kaufmann, Thomas F. Schulz. 8th edition. Springer, Berlin / Heidelberg 2016, ISBN 978-3-662-48677-1 , p. 479 ff .
- ^ Arnold S. Monto, Peter DeJonge, Amy P. Callear, Latifa A. Bazzi, Skylar Capriola, Ryan E. Malosh, Emily T. Martin, Joshua G. Petrie: Coronavirus occurrence and transmission over 8 years in the HIVE cohort of households in Michigan . In: Journal of Infectious Diseases. April 4, 2020, doi: 10.1093 / infdis / jiaa161 .
- ↑ Common coronaviruses are highly seasonal, with most cases peaking in winter months . on: ScienceDaily of April 7, 2020, Source: University of Michigan
- ↑ Common Human Coronaviruses are Sharply Seasonal, Study Says . on: Sci-News from April 8, 2020.
- ^ BW Neuman, MJ Buchmeier: Coronaviruses . Chapter 1 only: Supramolecular Architecture of the Coronavirus Particle . In: Advances in Virus Research . 1st edition. tape 96 . Elsevier Inc., 2016, ISBN 978-0-12-804736-1 , ISSN 0065-3527 , Section 2: Virion Structure and Durability, p. 4 + 5 , doi : 10.1016 / bs.aivir.2016.08.005 (English, full text [PDF; 1.3 MB ; accessed on June 15, 2020]).
- ↑ Yi Fan, Kai Zhao, Zheng-Li Shi and Peng Zhou: Bat Coronaviruses in China . In: Viruses . tape 11 , no. 3 . MDPI, March 2, 2019, p. 210 , doi : 10.3390 / v11030210 , PMID 30832341 , PMC 6466186 (free full text) - (English, full text [PDF; 0 kB ; accessed on June 16, 2020]).
- ↑ NCBI Taxonomy browser: Pacific salmon nidovirus . In: NCBI homepage . National Center for Biotechnology Information (NCBI), accessed May 21, 2020 .
- ↑ NCBI: Pacific salmon nidovirus (species)
- ↑ Gideon J Mordecai, Kristina M Miller, Emiliano Di Cicco, Angela D Schulze, Karia H Kaukinen, Tobi J Ming, Shaorong Li, Amy Tabata, Amy Teffer, David A Patterson, Hugh W Ferguson, Curtis A Suttle: Endangered wild salmon infected by newly discovered viruses . Version 1. In: eLife . eLife Sciences Publications Ltd., September 3, 2019, 47615, doi : 10.7554 / eLife.47615 (English, full text [PDF; 2.7 MB ; accessed on May 21, 2020]): “The novel nidovirus, named Pacific salmon nidovirus (PsNV), is most closely related to the recently described Microhyla alphaletovirus 1 […]."