Nucleic acids
Nucleic acids , also called nucleic acids , are macromolecules made up of individual building blocks, the nucleotides , which contain the genetic information of all organisms . Alternating simple sugars and phosphoric acid esters form a chain, with a nucleic base attached to each sugar . In addition to proteins , carbohydrates and lipids, nucleic acids form the fourth large group of biomolecules . Their most well-known representative as the basic type of nucleic acids is deoxyribonucleic acid (DNA or DNA); in all living beings this is the store of genetic information , only with some viruses ribonucleic acid (RNS or RNA) occurs instead in this function. In addition to their function as information stores, the nucleic acids, considered to be the “key molecules of life”, can also serve as signal transmitters or catalyze biochemical reactions.
history
The nucleic acid was first described by the Swiss physician Friedrich Miescher in 1869 after his examinations in the laboratory of the former kitchen of the Tübingen Castle . He worked for the founder of biochemistry , Felix Hoppe-Seyler . After Miescher gave up his research on proteins because they were too complex and diverse, he turned to the study of cell nuclei . Their function was completely unknown at the time. From the nuclei of white blood cells , he isolated a substance that differed significantly from proteins because of its high phosphorus content . He named it Nuclein from the Latin word nucleus (core). Although Miescher came very close to the function of Nuclein , he ultimately did not believe that a single substance could be responsible for inheritance.
"If we (...) wanted to assume that a single substance (...) is in some way (...) the specific cause of fertilization, one would undoubtedly have to think primarily of the nucleus."
In 1885 Albrecht Kossel announced that a nitrogen-rich base with the molecular formula C 5 H 5 N 5 had been isolated from a large amount of bovine pancreas , for which he suggested the name adenine , derived from the Greek word “aden” for gland . In 1889 Richard Altmann isolated an organic acid containing phosphorus, which he called nucleic acid, from the nucleus in addition to a protein-like component . In 1891 Kossel was able to produce yeast nucleic acid (using Altmann's method) and detect adenine and guanine as cleavage products. It turned out that a carbohydrate also had to be part of the nucleic acid. Kossel chose the name nucleic bases for the basic substances guanine and adenine and their derivatives . In 1893 Kossel reported that he had extracted nucleic acid from the calf's thymus glands and obtained a well-crystallized cleavage product, for which he proposed the name thymine . In 1894 he isolated another (basic) substance from the thymus glands. Kossel named this nucleic base cytosine .
After the structural formulas of guanine and adenine as the purine body and of thymine as the pyrimidine body had been finally elucidated at the end of the 19th century - mainly through the syntheses of Emil Fischer - Kossel and Hermann Steudel were also able to determine the structural formula of the nucleic base cytosine as a pyrimidine body without any doubt. It had meanwhile been shown that guanine, adenine, thymine and cytosine can be found in all viable cells.
The knowledge about these four nucleobases should be of essential importance for the later structure elucidation of the DNA. It was Albrecht Kossel who - together with a carbohydrate and phosphoric acid - clearly characterized them as building blocks of nucleic acid:
“I managed to get a number of fragments […] which are characterized by a very peculiar collection of nitrogen atoms. There are next to each other here [...] the cytosine, the thymine, the adenine and the guanine. "
Phoebus Levene proposed a chain-like structure of the nucleic acid. He coined the term “ nucleotide ” for the building blocks of nucleic acid. In 1929 he was able to identify the sugar content of the "animal" nucleic acid as deoxyribose . In the following it was referred to as deoxyribonucleic acid . It was recognized that deoxyribonucleic acid also occurs in plant cell nuclei.
In 1944, Oswald Avery , Colin McLeod and Maclyn McCarty were able to prove that nucleic acids are the stores of genetic information and not - as previously assumed - proteins.
The American James Watson (* 1928) and the Englishmen Francis Crick (1916-2004), Rosalind Franklin (1920-1958) and Maurice Wilkins (1916-2004) finally succeeded in clearing up the structure of deoxyribonucleic acid. Watson, Crick and Wilkins received the Nobel Prize in 1962 .
In 1977 Frederick Sanger , as well as Allan Maxam and Walter Gilbert , independently of one another, developed methods with which the order of the nucleotide building blocks, the sequence , could be determined. The chain termination method is now used in automated processes to sequence DNA.
construction
Chemical structure
Nucleic acids are chains with nucleotides as links. The central part of a nucleotide is the ring-shaped sugar molecule (in the picture in gray: the ribose). If the carbon atoms of this sugar are numbered clockwise from 1 to 5, a nucleic base (Fig. 1: red, green, yellow and blue) is attached to C1 via a glycosidic bond . At C3, a phosphate residue of the following nucleotide (blue) has formed an ester bond with the OH group of the sugar . A phosphate residue is also bound to the C5 of the sugar via the other of the two phosphodiester bonds .
In its unbound state, phosphoric acid has three acidic hydrogen atoms (on the OH groups) that can be split off. In a nucleic acid, two of the three OH groups are esterified and can therefore no longer release any protons. The third unbound acid function is responsible for the acidic character that gave nucleic acid its name. It can act as a proton donor or it is deprotonated in the cell (negative charge on the oxygen atom). Under physiological conditions ( pH 7), the nucleic acid is a large anion overall due to this negatively charged oxygen atom . When separating nucleic acids according to their size, an electric field can therefore be used, in which nucleic acids generally migrate to the anode (see agarose gel electrophoresis ).
The chains of nucleic acids are usually unbranched (either linear or closed in a ring, i.e., circular). For exceptions, see for example the Okazaki fragment , Holliday structure and clover leaf structure .
orientation
Their structure gives the nucleic acid a polarity or orientation in the chain building block sequence . It has a 5 'end (read: 5-line end), named after the C5 atom of the sugar to which a phosphate residue is bound, and a 3' end to which the free OH group at C3 Atom completes the chain. Usually, one writes down sequences, i.e. nucleotide sequences, starting with the 5 'end towards the 3' end. Polarity is very important in organisms. For example, there are DNA polymerases that can only build up a DNA strand in the 5 '→ 3' direction, and still others correct incorrectly incorporated nucleotides only in the 3 '→ 5' direction.
Spatial structure
In nucleic acids, the secondary structure is the spatial alignment. While the primary structure (the sequence) stores the information, the secondary structure determines the size, durability and also access to the stored information.
The simplest spatial structure is the double strand. Here two nucleic acid chains are facing each other in opposite directions. They are connected to one another via hydrogen bonds between the nucleobases. A pyrimidine base is paired with a purine base , the type of pair determining the stability of the double strand. Three hydrogen bonds are formed between guanine and cytosine , while adenine and thymine are only connected by two hydrogen bonds (see Figure 2). The higher the GC content (proportion of guanine-cytosine pairs), the more stable the double strand and the more energy (heat) has to be expended to split it into single strands. A double strand can consist of two different nucleic acid molecules or just a single molecule. At the end of the double strand, a loop is formed in which the chain "reverses" so that the opposite orientation arises.
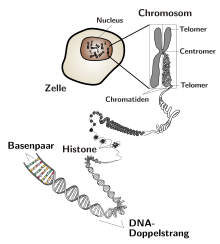
In DNA, as a result of the many different bond angles , the double strand winds around its own axis and forms a double helix . There are both left and right handed helices. This double strand, which is wound around itself, can then be twisted even further and wrap around other structures such as histones (special proteins). The point of this further tangling is to save space. Untwisted and stretched out, the DNA of a single human chromosome would be about 4 cm long.
Natural nucleic acids
Nucleic acids are found in all living organisms. Their task is, among other things, to save the genetic information, the blueprint of the respective organism, to exchange it with others of its kind and to pass it on to subsequent generations. In all organisms this is what DNA does. Only some viruses ( retroviruses such as HIV ) use the less stable RNA as a storage medium. However, hypothetical ribocytes, as the precursors of today's cellular organisms, could also have had an RNA genome in the primeval times of the earth ( RNA world hypothesis), but there has not yet been any evidence of this. Other nucleic acids as predecessors of RNA / DNA have also been discussed (XNA, see below).
Deoxyribonucleic acid (DNA, DNA)
DNA has deoxyribose as a sugar component (hence the name deoxyribo nucleic acid), which differs from ribose only in the lack of OH group on the C2 atom. The reduction of the OH group to a simple H only takes place at the end of the nucleotide synthesis. Deoxyribonucleotides thus arise from the ribonucleotides, the RNA building blocks. The difference, however, makes DNA chemically much more stable than RNA (justification see section RNA) and so stable that it can be detected dissolved in sea water (1 ppb) and estuaries (up to 44 ppb). The nucleobases adenine, cytosine, guanine and thymine occur in DNA, the latter being specific for DNA. Despite the small number of four different basic modules, a lot of information can be stored.
- Sample calculation:
- A piece of DNA made up of 4 possible basic building blocks with a total length of 10 base pairs gives 4 10 = 1,048,576 possible combinations
- The genome of the E. coli bacterium is approximately 4 × 10 6 base pairs. Since there are 4 possibilities (A, C, G or T) for a base pair, it corresponds to 2 bits (2 2 = 4). This means that the entire genome has an information content of 1 megabyte .
The DNA is in the form of a double strand that is wound around itself to form a double helix. Of the three helix types identified by X-ray structure analysis, only the B-DNA has so far been detected in vivo . It is a right-handed helix with a pitch (length of the helix for a complete turn) of 3.54 nm and 10 base pairs and a diameter of 2.37 nm. There is also the wider A-helix (pitch 2.53 nm; diameter 2 , 55 nm) and the more elongated Z-helix (pitch 4.56 nm; diameter 1.84 nm). If a gene encoded in the DNA is to be read or if the DNA itself is to be doubled in the course of cell division, the helix is untwisted on a part by enzymes ( topoisomerases ) and the double strand is split into single strands ( helicases ).
In bacteria , the DNA is present as a ring-shaped molecule, while in eukaryotes it has free ends, the so-called telomeres . The nature of the DNA replication mechanism means that linear DNA molecules are shortened by a few base pairs per duplication. The more often a cell divides, the shorter the DNA becomes. This has no consequences with limited cell division, since at the end of such a strand there are short sequences that are repeated several thousand times. So no genetic information is lost. The shortening is partly compensated for by the enzyme telomerase (only in stem cells and cancer cells). If the length of the repetitive sequences at the end of the strand falls below a certain length, the cell no longer divides. This is one of the reasons for a limited lifespan. Because bacteria have a circular DNA molecule, the strand does not shorten.
Ribonucleic acid (RNS, RNA)
The OH group on the C2 atom of the ribose is responsible for the lower stability of the RNA. This is because, like the OH group on the C3 atom for normal chain formation, it can form a link with the phosphate residue. If such a transesterification occurs spontaneously , the nucleic acid chain is interrupted.
 Cytosine |
 Uracil |
 Thymine |
Another difference is that thymine is used in DNA, while uracil is used in RNA . Nucleic bases within the DNA can be chemically changed by oxidative conditions or other influences. Occasionally, deamination occurs (splitting off of an NH 2 group, an O = group is formed instead). In a double strand, the sites for hydrogen bridges on the opposite nucleic bases no longer fit together and partial splitting occurs. Enzymes can cut out and replace or repair altered nucleobases. To do this, use the second, unchanged nucleobase as a template. If such a deamination occurs with cytosine, uracil is formed. If uracil were also commonly found in DNA, an enzyme could no longer distinguish whether the uracil is the wrong nucleobase or the opposite guanine (which was previously paired with cytosine). In this case, important information could be changed and a mutation could occur. In order to avoid this confusion, thymine is not used in the DNA. Uracil is recognized and removed in the DNA by specific enzymes, the uracil glycosylases. Enzymes can recognize thymine perfectly due to its additional methyl group and so it is clear that every uracil in the DNA is a broken cytosine. In RNA, this risk of information corruption is not serious, since information is only stored here for a short time and there are not just one RNA molecule of the respective type, but hundreds. If some of them are defective, this has no serious impact on the entire organism, as there are enough replacements.
Synthetic nucleic acids
There are numerous variants of the above standard nucleic acids RNA and DNA. Some of these are of natural origin, but variants have also been developed within the framework of xenobiology , the building blocks of which at first glance are no longer recognizable as ribo- (in the case of RNA ) or deoxyribonucleotides (in the case of DNA ). In individual cases it is still the subject of discussion to this day whether a certain variant occurs in nature (or, for example, occurred in the initial phase of life on earth) or not. In principle, all three parts of a nucleic acid building block can be changed, i.e.:
- the bases: In addition to naturally occurring non-standard bases (in the simplest case uracil for DNA) and modifications ( DNA methylation ), u. a. developed the following artificial variants :
- xDNA , xxDNA, yDNA, yyDNA (2006)
- Hachimoji DNA (2019)
- the sugars: xenonucleic acids (XNA) have another group instead of ribose or deoxyribose, which can, but does not have to be, a different sugar or sugar derivative. These are u. a .:
- Cyclohexene nucleic acids ( English cyclohexenyl nucleic acids , CeNA)
- Tricyclo-Deoxyribonucleic Acids (tcDNA)
- the phosphate group
- Phosphorothioate -deoxyribonucleic acid , see Fomivirsen
- N3′-P5′- phosphoramidates (NP)
- Morpholino-Phosphoramidate (MF)
- Deoxyadenosine mono arsenate (dAMAs) see GFAJ-1 §Discussion about the incorporation of arsenic into biomolecules : questionable incorporation into DNA in the Halomonas species GFAJ-1 (see also Halomonas titanicae )
- a combination of these and other special modifications:
- Peptide nucleic acid ( English Peptide Nucleic Acid , PNA , also PNS)
- bridged nucleic acid ( english locked nucleic acid , LNA) - in contrast to unbridged nucleic acid ( english unlocked nucleic acid , UNA)
- RNA occurs in living organisms as D RNA on - L RNA as so-called. Spiegelmer however, can be synthesized. The same applies analogously to DNA. L -DNA is broken down more slowly by enzymes than the natural form, which makes it interesting for pharmaceutical research .
See also
literature
- Hans Beyer , Wolfgang Walter : Textbook of organic chemistry. 23rd edition. Hirzel, Stuttgart 1998, ISBN 3-7776-0808-4 .
- Lubert Stryer : Biochemistry. 5th edition. Spektrum Verlag, 2003, ISBN 3-8274-1303-6 .
- Ralf Dahm: The molecule from the castle kitchen . In: MaxPlanckResearch . No. 1 , 2004, p. 50–55 ( mpg.de ( memento of April 11, 2011 in the Internet Archive ) [PDF; 1000 kB ; accessed on December 25, 2019]).
Individual evidence
- ↑ Entry on nucleic acids. In: Römpp Online . Georg Thieme Verlag, accessed on February 17, 2016.
- ↑ a b Ulrike Roll: nucleic acids. In: Werner E. Gerabek , Bernhard D. Haage, Gundolf Keil , Wolfgang Wegner (eds.): Enzyklopädie Medizingeschichte. De Gruyter, Berlin / New York 2005, ISBN 3-11-015714-4 , pp. 1060 f .; here: p. 1060.
- ↑ A. Kossel: About a new base from the animal body. Lecture in reports of the German Chemical Society. Issue 18, 1885, p. 79.
- ^ A. Kossel: Further contributions to the chemistry of the cell nucleus. In: Journal of Physiological Chemistry. Volume 10, 1886, p. 248.
- ^ R. Altmann: About nucleic acids. In: Archives for Anatomy and Physiology, Physiological Department. Leipzig 1889, pp. 524-536.
- ↑ A. Kossel: About the chemical composition of the cell. Lecture. In: Archive for Anatomy and Physiology / Physiological Department 1891. p. 178.
- ^ A. Kossel, A. Neumann: About thymine, a cleavage product of nucleic acid. In: Reports of the German Chemical Society. Volume 26, 1893, p. 2753; Preparation and cleavage products of nucleic acid (adenylic acid). Lecture. In: Reports of the German Chemical Society. Volume 27, 1894, p. 2215; About nucleic acid and thymic acid. In: Journal of Physiological Chemistry. Volume 22, 1896-97, p. 74.
- ↑ A. Kossel, H. Steudel: Further investigations on cytosine. In: Hoppe-Seyler's journal for physiological chemistry . Volume 38, 1903, p. 49.
- ↑ A. Kossel: About the chemical composition of the cell nucleus. Nobel Lecture on October 12, 1910 in Stockholm. In: Munich Medical Weekly . Volume 58, 1911, p. 65.
- ^ P. Levene, E. London: The structure of Thymonucleic acid. In: Journal of Biological Chemistry . 1929. 83. pp. 793-802.
- ↑ Philippine Aupy, Lucía Echevarría, Karima Relizani, Fedor Svinartchouk, Luis Garcia, Aurélie Goyenvalle et al. : Identifying and Avoiding tcDNA-ASO Sequence-Specific Toxicity for the Development of DMD Exon 51 Skipping Therapy , in: Molecular Therapy - Nucleic Acids Volume 19, pp. 371–383, March 6, 2020, online November 26, 2019, doi: 10.1016 / j.omtn.2019.11.020
- ↑ Pradeep S. Pallan, Damian Ittig, Annie Héroux, Zdzislaw Wawrzak, Christian J. Leumannb, Martin Egli: Crystal structure of tricyclo-DNA: an unusual compensatory change of two adjacent backbone torsion angles , in: Chemical Communications, Volume 7, 2008 , doi: 10.1039 / B716390H
- ↑ Damian Ittig, Anna-Barbara Gerber, Christian Joerg Leumann: Position-dependent effects on stability in tricyclo-DNA modified oligonucleotide duplexes , in: Nucleic Acids Research 39 (1), pp. 373-380, January 2011, doi: 10.1093 / nar / gkq733 , PMID 20719742 , PMC 3017593 (free full text)
- ^ W. Purschke, F. Radtke, F. Kleinjung, S. Klussmann: A DNA Spiegelmer to staphylococcal enterotoxin B , in: Nucleic Acids Research. Volume 31, No. 12, 2003, pp. 3027-3032, doi: 10.1093 / nar / gkg413 , PMID 12799428
- ↑ Gosuke Hayashi, Masaki Hagihara, Kazuhiko Nakatani: Application of L-DNA as a molecular tag , in: Nucleic Acids Symposium Series , Volume 49, No. & mbsp: 1, 2005, pp. 261–262, doi: 10.1093 / nass / 49.1.261 , PMID 17150733


