Selenocysteine
| Structural formula | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|

|
||||||||||||||||||||||
| Structure of the naturally occurring L -elenocysteine | ||||||||||||||||||||||
| General | ||||||||||||||||||||||
| Surname | Selenocysteine | |||||||||||||||||||||
| other names |
|
|||||||||||||||||||||
| Molecular formula | C 3 H 7 NO 2 Se | |||||||||||||||||||||
| External identifiers / databases | ||||||||||||||||||||||
|
||||||||||||||||||||||
| properties | ||||||||||||||||||||||
| Molar mass | 168.0 g mol −1 | |||||||||||||||||||||
| safety instructions | ||||||||||||||||||||||
|
||||||||||||||||||||||
| As far as possible and customary, SI units are used. Unless otherwise noted, the data given apply to standard conditions . | ||||||||||||||||||||||
Selenocysteine is an amino acid. L -elenocysteine (abbr. Sec or U ) is the 21st proteinogenic L - amino acid and a reactive analogue of the natural L - cysteine . Selenocysteine contains a selenium atom instead of the sulfur atom .
Isomerism
Selenocysteine can exist in the enantiomeric forms D and L , with proteins only in the L form, also synonymous with ( R ) -selenocysteine. D -elenocysteine is enantiomeric to L -elenocysteine and is of little importance; In the scientific literature, “selenocysteine” (without a prefix) always stands for L- selenocysteine.
| Isomers of selenocysteine | ||
| Surname | L -elenocysteine | D -elenocysteine |
| other names | ( R ) selenocysteine | ( S ) selenocysteine |
| Structural formula | 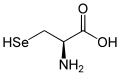 |

|
| CAS number | 10236-58-5 | 176300-66-6 |
| 3614-08-2 ( DL ) | ||
| EC number | 808-428-7 | - |
| - ( DL ) | ||
| ECHA info card | 100.236.386 | - |
| - ( DL ) | ||
| PubChem | 25076 | 5460539 |
| 6326972 ( DL ) | ||
| Wikidata | Q408663 | Q27110363 |
| Q65017088 ( DL ) | ||
properties
L -Selenocystein is with the amino acid L related chemically closely -cysteine, but has a lower acidity constant pK s = 5.3 for the selenol in comparison with pK s = 8-10 for the thiol group of the L -cysteine. Selenocysteine is also more redox-active than cysteine. These properties are likely to be a major reason for the incorporation of L -selenocysteine into enzymes .
Selenocysteine is mainly present as an inner salt or zwitterion , the formation of which can be explained by the fact that the proton is split off from the carboxy group and taken up by the lone pair of electrons on the nitrogen atom of the amino group .
The zwitterion does not migrate in the electric field because it is uncharged as a whole. Strictly speaking, this is the case at the isoelectric point (at a certain pH value), at which the selenocysteine also has its lowest solubility in water.
biochemistry
The genetic code applies to all forms of life, but there are special features. While the standard code enables cells to produce proteins from the 20 canonical α- amino acids , representatives of the archaea , bacteria and eukaryotes can incorporate selenocysteine during translation via a mechanism known as recoding . The incorporation of L- selenocysteine as an additional proteinogenic amino acid often only enables the functionality of some essential enzymes.
Occurrence
Since humans, many animals, some algae and protozoa lacks the codon for the vital 21st amino acid selenocysteine, the stop codon UGA is read out as Sec, which is a complex process. Even in insects or nematodes, some representatives form selenocysteine, others do not. For a long time, fungi were considered to be organisms completely lacking selenocysteine, until nine species were found among around 1000 species that produce selenocysteine.
More than 30 eukaryotic and more than 15 bacterial selenocysteine-containing proteins are known. So one found in mammals u. a. various glutathione - peroxidases , tetraiodothyronine - deiodinases or large thioredoxin reductases and in bacteria and archaea formate dehydrogenases, hydrogenases, protein components of the glycine reductase and D - proline reductase systems and several enzymes of the metabolic pathway of methane formation as selenocysteine-containing enzymes .
Many of the enzymes mediate redox reactions . With them the reactive selenocysteine is in the active center . Glutathione peroxidase is important for eukaryotes as part of the cellular defense against damage caused by oxidative stress . The function of the selenoproteins is disturbed when there is a selenium deficiency. For example, Keshan's disease - a cardiomyopathy associated with Coxsackie virus infections - occurs more frequently when there is a lack of the trace element selenium in the diet; the Kashin-Beck disease comes as a syndrome in areas with selenium-poor soils before. However, the ( SELECT ) study did not confirm the assumption that dietary supplements containing selenium generally prevent cancer .
biosynthesis

L -elenocysteine is produced biosynthetically by selenylation of an L -serine, which is bound to a specific tRNA :
- Binding of the α-amino acid L - serine ( Ser ) to a special tRNA, the tRNA Sec with the anticodon UCA (5 ′ → 3 ′ noted).
- This Ser-tRNA Sec is selenylated , i. H. the L -serine is converted to L -elenocysteine ( Sec ) by replacing the hydroxy group of the side chain with selenol ( SeH ). This creates the Sec-tRNA Sec .
The biosynthetic pathway differs significantly from other amino acids, which are initially formed as free amino acids and only then bound to a tRNA.
Recoding
The tRNA Sec has the anticodon UCA and this triplet, 3'-ACU-5 ' in the opposite direction , can pair with the base triplet of the codon of UGA the mRNA . Normally causes a stop codon , the termination of translation. However, if special sequences of the mRNA form a hairpin structure , it becomes possible to pair the loaded Sec-tRNA Sec with the codon . The stop signal is thus ignored and selenocysteine is incorporated into the protein at this point. This process is also known as recoding .
UGAUGA
In bacteria, such a place SECIS ( se leno c ysteine i nsertion s equence ) said sequence of the mRNA immediately adjacent to the codon UGAthen recoded and only this will be adjacent. The Secis sequence is recognized by a specific GTP- dependent translation factor , the elongation factor SelB , which at the same time binds the Sec-tRNA Sec . After the selenocysteine has been incorporated, the Secis sequence is also read from the ribosome and translated into the corresponding amino acids of the protein.

In eukaryotes and archaea, on the other hand , a Secis sequence is attached to the 3 'end of the mRNA, is not read from the ribosome, and allows all codons of UGAthis mRNA to be recoded. For example, the human selenoprotein P contains selenocysteines in ten positions.
The incorporation of L- selenocysteine during protein synthesis in eukaryotes requires additional factors (see figure):
- The Sec-tRNA Sec is bound by the specific GTP-dependent translation factor mSelB .
- mSelB forms a complex with another protein SBP2 , which recognizes the Secis sequence and binds to it.
- This complex (shown in the figure as a green ball) enables the reinterpretation: If a codon is
UGAread on the ribosome and paired with the anticodon of the Sec-tRNA Sec , then selenocysteine is now incorporated at this position. - With this type of recoding, the translation must then be terminated by another stop codon , either
UAAorUAG( marked as stop in the figure ).
The conditions in the selenoprotein synthesis of the archaea have not yet been clarified.
literature
- Joseph W. Lengeler, G. Drews, Hans Günter Schlegel: Biology of the prokaryotes . Thieme, Stuttgart 1999, ISBN 3-13-108411-1 , p. 185 ff .
Web links
- Translation process - incorporation of Sec
- Selenocysteine - some more details (pdf, in English; 324 kB, 20 pages) - Errata: On page 19, every SeCys has an Se (selenium) instead of an S (sulfur) as shown, with Cys itself S is correct.
Individual evidence
- ↑ There is not yet a harmonized classification for this substance . A label of [No public or meaningful name is available] in the Classification and Labeling Inventory of the European Chemicals Agency (ECHA), accessed on July 10, 2019, is derived from a self-classification by the distributor .
- ↑ Marco Mariotti, Gustavo Salinas, Toni Gabaldón, Vadim N. Gladyshev: Utilization of selenocysteine in early-branching fungal phyla . In: Nature Microbiology . February 11, 2019. doi : 10.1038 / s41564-018-0354-9 .
- ↑ Jun Lu, Arne Holmgren: Selenoproteins . In: Journal of Biological Chemistry . Volume 284, No. 2, January 2009, pp. 723-727. doi : 10.1074 / jbc.R800045200 . PMID 18757362 .
- ↑ M. Reeves, P. Hoffmann: The human selenoproteome: recent insights into functions and regulation . In: Cellular and Molecular Life Sciences . Volume 66, No. 15, August 2009, pp. 2457-2478. PMID 19399585 . PMC 2866081 (free full text).
- ↑ Berry, MJ, Banu, L., Harney, JW, Larsen, PR: Functional Characterization of the Eukaryotic SECIS Elements which Direct Selenocysteine Insertion at UGA Codons . In: The EMBO Journal . 12, No. 8, 1993, pp. 3315-3322. PMID 8344267 . PMC 413599 (free full text).
- ↑ Burk RF, Hill KE: Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis . In: Annu Rev Nutr . 25, 2005, pp. 215-235. doi : 10.1146 / annurev.nutr.24.012003.132120 . PMID 16011466 .
- ↑ Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A: Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation . In: The EMBO Journal . 1, 2000, pp. 4796-4805. doi : 10.1093 / emboj / 19.17.4796 . PMID 10970870 .

