Hepatitis C virus
| Hepatitis C virus | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
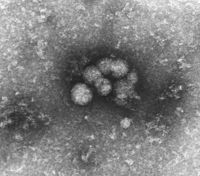
HCV virions from serum |
||||||||||||||||||
| Systematics | ||||||||||||||||||
|
||||||||||||||||||
| Taxonomic characteristics | ||||||||||||||||||
|
||||||||||||||||||
| Scientific name | ||||||||||||||||||
| Hepacivirus C | ||||||||||||||||||
| Short name | ||||||||||||||||||
| HCV | ||||||||||||||||||
| Left | ||||||||||||||||||
|
The Hepatitis C virus is an enveloped , single-stranded RNA virus with positive polarity (ss (+) RNA ), and is the causative agent of hepatitis C . It is the only known RNA virus that (without being a retrovirus ) can cause a chronic infectious disease. At diagnosis , disease and prevention see Hepatitis C .
Although an infectious agent had been known to cause what was then called “Non-A-Non-B hepatitis” since the first reports in 1974, the virus was not identified for a long time. It was only after the first application of cloning genome fragments from the serum of a chimpanzee artificially infected with HCV that the pathogen, then known as hepatitis C virus, was discovered in 1989 and sequenced in 1990 . It is also the first virus whose genome sequence and virus proteins are subject to comprehensive patent protection, which made the development of new test methods considerably more difficult. Following the takeover of Chiron Corporation in 2006, the patent holder is the pharmaceutical company Novartis .
The hepatitis C virus (HCV) belongs together with the hepatitis B virus (HBV), the Epstein-Barr virus (EBV), the human papillomavirus (HPV), human T-lymphotropic virus 1 (HTLV-1) and the human herpesvirus 8 (HHV-8, also Kaposi's sarcoma herpesvirus, KSHV) to the human viruses that can cause cancer. It is estimated that these viruses are responsible for 10 to 15 percent of all cancers worldwide .
Scientists Harvey J. Alter , Michael Houghton and Charles M. Rice were awarded the Nobel Prize in Physiology or Medicine in 2020 for the discovery of the virus .
Molecular biology
Systematics
Through sequence comparisons and the homology to other virus proteins, the HCV was grouped into the Flaviviridae family , although initially the classification into the genera Flavivirus or Pestivirus was not clear. Phylogenetically, the HCV seems to be more closely related to the otherwise only animal pathogenic pestiviruses, this is suggested by the hydrophobicity profiles of the proteins as well as the genome organization . It seemed taxonomically sensible to group the HCV as the only representative in a new genus Hepacivirus . The GB viruses (GB virus A and B in New World monkeys and tamarins ), especially GB virus C , which also occurs in humans but is non-pathogenic, are closely related to HCV, but not yet assigned to a genus or other genera .
Variability and subtypes
The variability of the HCV is also relatively high compared to other single-stranded RNA viruses. At the genome level, when comparing different isolates, up to 40% deviations can be found. Therefore, the HCV is divided into seven genotypes (1-7), whereby a uniform genotype must have at least 72% agreement at the amino acid level. These genotypes are further subdivided into 30 subtypes (1a, 1b, 3a etc.), the homology at the amino acid level being 73 to 86% within the subtypes.
Due to the reading inaccuracy of the viral RNA polymerase with a mutation rate of up to per nucleotide and replication, further deviations from the original sequence, which are referred to as quasi-species , develop in an infected host . The rapid emergence of this quasi-species has been linked to the ability of HCV to cause chronic infection. The constantly appearing variants (similar to the HIV infection) avoid the access of the immune system; this mechanism is also called immune evasion .
structure
The single-stranded RNA genome (negatively charged as acid) is attached to the core protein (basic, positively charged) in the virion . The core protein most likely does not form a classic capsid with a fixed symmetry (e.g. icosahedral). So far, free capsids could not be credibly depicted or stored together.
The core protein is from the inside in the viral envelope anchored and a transmembrane helix to the complex of the two envelope proteins E1 / E2 ( envelope engl. Sheath) attached. The transmembrane segment of the HCV core protein probably corresponds to the (larger) M (matrix) protein in viruses of the genus Flavivirus (e.g. yellow fever virus , TBE virus ). This M protein is split off from the core protein and lines the virus envelope from the inside; in the case of HCV this cleavage of an M protein apparently does not take place.
The HCV is extremely difficult to visualize in an electron microscope , so far no publication has been able to show a finally proven picture. Some images show particles with an assumed size of approx. 50 nm, which become visible after certain cleaning processes (see above). This difficult visualization, probably due to structural instability and sensitivity to normal preparation methods of HCV, is a reason for its very late discovery despite intensive search.
In the genome of HCV is a major place of open reading frames ( open reading frame , ORF) from which a single polyprotein 3008-3037 amino acids in length is read. This protein is split into structural proteins (core, E1 and E2) and non-structural proteins (NS2 to NS5) by cellular proteases ( signalase ) of the ER membrane and viral proteases during translation . This means that apart from the virus genome present, no further messenger RNA is transcribed. The table lists these proteins with their (partly still unclear) function, molar mass and amino acid position on the polyprotein:
| protein | Size ( kDa ) | position | function |
|---|---|---|---|
| Core | 21 (19) | 1-190 | Binding of the RNA and the E1 / E2 heterodimer
Oligomerization and lipid binding , activation and inhibition of cellular genes ( leucine zipper motif) |
| E1 | 31-37 | 191-383 | Glycosylated membrane protein , viral coat protein, E1 / E2 heterodimerization |
| E2 | 61-72 | 384-746 | Glycosylated membrane protein, viral coat protein, binding to cellular receptors (CD81, SRB1),
Hypervariable Region (HVR1), antibody binding region |
| p7 | 7th | 747-809 | Membrane protein, possibly ion channel |
| NS2 | 21-23 | 810-1026 | Zinc metalloprotease , autoprotease in connection with NS3 |
| NS3 | 70-72 | 1027-1657 | Protease , helicase |
| NS4A | 16-27 | 1658-1710 | Membrane association of the NS3-NS4A complexes, cofactor for NS3 |
| NS4B | 27 | 1711-1971 | membrane-associated, unknown function |
| NS5A | 56-58 | 1972-2419 | Phosphoprotein, previously associated with interferon sensitivity |
| NS5B | 68-70 | 2420-3010 | RNA-dependent RNA polymerase |
The open reading frame of about 9.5 kb long HCV genome is of two non-coding regions (NCR, engl. Non-coding region ) flanked which play a regulatory role during viral replication. In front of the start codon (defined as nucleotide position 1) is the approximately 340 base long 5 'NCR, which shows the greatest sequence correspondence and a complex folding of the RNA strand within the HCV isolates. At the end of the genome is the 250-300 base long 3'-NCR. This essentially consists of a polymeric uracil (polyU) or adenine ( polyA ) and a highly conserved nucleotide sequence of 98 bases in length, which is known as the X-tail .
Replication
The recognition of the liver cell ( hepatocyte ) as a target cell and entry into the cell is very likely mediated via one or more receptors . The receptor for HCV has not yet been clearly identified; some evidence suggests binding to the beta- lipoprotein receptor, the CD81 receptor and the HDL receptor (SRB1). By inhibiting CD81 and SRB1, the uptake of E1 / E2 in hepatocytes could be blocked, but none of these receptors alone is sufficient to infect a cell. In addition, none of the mentioned receptors is liver-specific.
In analogy to other members of the Flaviviridae , it is assumed that the HCV enters the cell through a constricting vesicle of the cell membrane ( endosome ), where it fuses the endosome membrane with the virus envelope and thereby releases the core protein-RNA complex into the Cytosol is coming. The HCV genome, which can be read as mRNA, first reaches the ribosomes of the rough ER , where the first synthesis of virus proteins takes place. This first step is necessary because the virus first needs some molecules of the viral RNA-dependent RNA polymerase NS5B and this enzyme is not present in the cell.
In order to increase the amount of available viral mRNA, the next step is the replication of the viral RNA. For this purpose, the NS5B polymerase synthesizes a counter-strand (negative strand) of the viral (+) RNA as a template (template) for the subsequent synthesis of further plus strands. The specifically folded sections in the two non-coding regions of the viral RNA probably serve as the initiation signal for the NS5B polymerase. There are indications that both ends of the RNA genome close to form a ring by binding to cellular proteins, thereby creating a replication complex in the sense of a rolling circle mechanism . In HCV-replicating cell cultures, conspicuous membrane structures could be found under the electron microscope, which are regarded as HCV replication complexes. These structures, known as “ membranous web ” (membranous network), are induced by the hydrophobic membrane protein NS4B. Very similar specific membrane changes are found as so-called viroplasm in all (+) ss-RNA viruses that were examined in this regard.
After the viral mRNA has been sufficiently synthesized, translation of the viral proteins, predominantly the structural proteins, begins . To initiate translation on the ribosomes, eukaryotic mRNAs usually use a modification of the 5 'end, the so-called cap . However, the HCV has by the folding of the 5'-NCR a special structure for cap-independent initiation, the so-called internal ribosomal entry site (engl. Internal ribosomal entry site , IRES). As a result, the HCV RNA does not require any cellular factors to bind to the ribosomes and to start protein synthesis. The IRES is usually only found in some picornaviruses such as the poliovirus .
Even during translation, the structural proteins are cut off by cellular proteases ( signalase ) from the resulting polyprotein strand; the envelope proteins E1 and E2 reach the lumen of the ER, store themselves in the membrane and are glycosylated. The core protein is stored externally in the ER membrane and, due to its charging properties, binds the viral mRNA. Now it comes to the packaging of the core RNA complex and the constriction (budding, engl. Budding ) of the HCV particle in the lumen of the ER. The new virions now leave the cell through cellular secretion via the Golgi apparatus .
Hosts
Humans are the only natural hosts of the HCV. In the study of HCV in the 1980s and attempts to prepare a vaccine from 1990 were apes (mainly chimpanzees ) infected artificially. These can also be infected, but chronic infection is rare. These tests, which were carried out mainly in the USA at the time, are prohibited in Europe for ethical reasons.
Despite intensive experiments, a virus-producing cell culture system for HCV has only been available since 2004, which should significantly accelerate research into the replication of the virus in cells and the development of new antiviral substances.
Epidemiology
The prevalence ratio (number of sick people in relation to the number of people examined) is 0.031 worldwide, in Europe and the USA less than 0.02. In Japan, Mongolia and Egypt it is 0.181, in the latter caused by errors in treatment for schistosomiasis (contaminated cannulas). The WHO assumes 170 million chronic virus carriers worldwide. The number of people with hepatitis C is much lower in some countries.
The distribution of the genotypes and subtypes also follows geographical patterns: in Europe and America, the subtypes 1a, 1b and 3a (mainly among drug addicts), which occur worldwide, are predominantly found; in Asia, subtype 1b is dominant, in Africa genotype 4, in South Africa genotype 5 and in Hong Kong and Vietnam genotype 6. Genotypes 2 and 3 are represented worldwide, but to a much lesser extent.
The period of the genotypic separation of the genotypes could be estimated at around 500 years using the sequence variability and the calculated mutation frequency. It can therefore be assumed that the diversification of genotypes went hand in hand with the advent of global seafaring around the year 1500.
transmission
The virus is transmitted parenterally ; H. primarily through blood and blood products; a sexual transmission is rare. This defines the following risks: dialysis (especially before 1991), intravenous drug use , tattooing and piercing . The transmission route is unknown in around 30% of patients.
Antivirals
Approved and experimental antiviral agents against the hepatitis C virus include Ribavirin against the RdRP , the NS3 / 4A inhibitors such asunaprevir , boceprevir , Ciluprevir , Danoprevir , Faldaprevir , Glecaprevir , Grazoprevir , Narlaprevir , Paritaprevir , Simeprevir , Sovaprevir , telaprevir , Vaniprevir , Vedroprevir , voxilaprevir and the NS5A inhibitors daclatasvir , elbasvir (approved for HCV1 and HCV4), ledipasvir , MK-8408 , odalasvir , ombitasvir , ravidasvir and velpatasvir .
Reporting requirement
In Germany, all direct or indirect evidence of the hepatitis C virus must be reported by name in accordance with Section 7 of the Infection Protection Act .
In Switzerland, the positive laboratory analytical finding is a hepatitis C virus notifiable and that after the Epidemics Act (EpG) in connection with the epidemic Regulation and Annex 3 of the Regulation of EDI on the reporting of observations of communicable diseases of man .
sources
- H.-J. Thiel et al: Genus Hepacivirus. In: CM Fauquet, MA Mayo et al: Eighth Report of the International Committee on Taxonomy of Viruses . London, San Diego 2005, ISBN 0-12-249951-4 , pp. 993-998.
- David M. Knipe, Peter M. Howley (eds.-in-chief): Fields' Virology. 5th edition. 2 volumes. Philadelphia 2007, ISBN 978-0-7817-6060-7 , pp. 1113-1126 and 1253-1291.
Web links
- Hepatitis C virus (taxonomy database of the NCBI)
- Reference sequence NC 004102 of the HCV
Individual evidence
- ↑ ICTV Master Species List 2018b.v2 . MSL # 34, March 2019
- ↑ a b c d ICTV: ICTV Taxonomy history: Yellow fever virus , EC 51, Berlin, Germany, July 2019; Email ratification March 2020 (MSL # 35)
- ↑ D. Martin, JS Gutkind: Human tumor-associated viruses and new insights into the molecular mechanisms of cancer . In: Oncogene . tape 27 , no. 2 , 2008, p. 31-42 , PMID 19956178 .
- ^ The Nobel Prize in Physiology or Medicine 2020 . In: nobelprize.org, October 5, 2020).

