Influenza A virus H1N1
| Influenza A virus H1N1 | ||||||||
|---|---|---|---|---|---|---|---|---|
TEM image of H1N1 influenza A viruses |
||||||||
| Systematics | ||||||||
| Taxonomic characteristics | ||||||||
|
||||||||
| Scientific name | ||||||||
| Influenza A virus A / H1N1 | ||||||||
| Short name | ||||||||
| FLUAV / (H1N1) | ||||||||
| Left | ||||||||
|
Influenza A virus H1N1 (A / H1N1) denotes a subtype of the influenza A virus (genus Alphainfluenzavirus ) from the family of Orthomyxoviruses ( Orthomyxoviridae ), which occurs in ducks , humans and pigs , but also infects numerous other mammalian species and turkeys can. In humans and pigs, H1N1 influenza viruses became established as a result of the Spanish flu of 1918. This subtype has caused tens of millions of human deaths. In the spring of 2009, a previously unknown subtype of the H1N1 virus, the so-called swine flu virus, spread in North America and caused another pandemic ( pandemic H1N1 2009/10 ).
The first human outbreak: the Spanish flu
The Spanish flu of 1918 showed several peculiarities: With this H1N1, a new influenza subtype was alleged to have appeared. The first three waves followed each other within a few months, which is very unusual, and it was only with the third wave that they adhered to the "influenza season" in late winter, which is common in temperate latitudes. The mortality focused on the age group of 15 to 40 years of age; Pregnant women in the last third of their pregnancy turned out to be extremely at risk . Although the data situation is contradictory on this point, the causative agent of the Spanish flu also seems to have been more infectious than the usual influenza viruses. The epidemic resulted in very different mortality rates in different regions of the world, with countries in tropical and subtropical climates generally being particularly hard hit.
The Spanish flu was the largest epidemic of modern times , although the exact number of deaths remains unknown. Most of the deaths were not from the virus directly, but from secondary bacterial pneumonia . For decades the death toll was estimated at 21 million, which seems too low given that 17 to 18 million people died in British India alone ; this information is well supported by population censuses. An even remotely accurate balance sheet cannot be drawn because civil war raged in two densely populated states - the former tsarist empire and China . An extrapolation for these two countries on the basis of the Indian figures is dubious because of the very different conditions there.
The circumstances of the outbreak are unclear. The first safe news relates to soldiers who were prepared for service in the First World War in training camps in the USA in March 1918 . From the United States, Japan, the Philippines, Canada, and Mexico were infected; American soldiers brought the disease to the Western Front in France, from where it spread to Spain and South America, the rest of Europe, Africa and Asia. Because international press agencies first reported the epidemic from Spain, its origin was wrongly attributed to this country.
Water birds are the original reservoir of influenza viruses. Jeffery Taubenberger , who later reconstructed the pathogen causing the Spanish flu, says that shortly before 1918 a complete influenza virus passed from birds to humans. Other researchers contradict him and suspect a recombination (English: reassortment ) with gene segments from swine influenza viruses. At the time of the Spanish flu, pig disease was also reported in Harbin , China and Iowa in the United States, but at least in those cases the sequence appears as if they had been infected by humans. Genetic studies on the hemagglutinin gene also suggest this process.
It is controversial whether the H1N1 virus actually circulated among humans for the first time in 1918. One can also interpret the relative protection of the over 45-year-olds in the Spanish flu in such a way that they were already exposed to H1N1 viruses in the period before 1873 and thus immune.
With the third wave in the winter of 1918/19, the Spanish flu rejoined the usual influenza season in the temperate latitudes. But the fourth wave in the winter of 1919/20 showed - albeit to a lesser extent - the characteristic increased mortality among young adults. As a result of the disease, stable lines of H1N1 viruses established themselves among humans and pigs. It can be assumed that influenza viruses of the subtype H1N1 have circulated in mankind since 1918, due to virus isolates from that year. Serious influenza epidemics , which affected at least Europe and North America, still occurred in 1928/29, 1932/33, 1936/37 and 1943/44.
The discovery of the virus

The pathogen was still unknown during the Spanish flu. Some researchers had already suspected a "filterable virus" (where "virus" initially meant "pathogen" or even "poison"). In the textbooks, however, the bacterium that is now called Haemophilus influenzae was considered to be the cause of influenza. With the usual methods of bacteriology , however, no pathogen could be detected: it could neither be stained nor grown on artificial culture media. If there has been an advance in knowledge through the Spanish flu, it is to the extent that the actual pathogen of this flu variant was still unknown at the time, i.e. Haemophilus influenzae was eliminated as a pathogen.
Classic swine influenza has long been confined to the northern and midwestern United States, where annual winter outbreaks occurred after 1918. In 1930 Richard Shope isolated an influenza virus from pigs for the first time at the Rockefeller Institute in Princeton. Two years later, the British researchers Wilson Smith , Christopher Andrewes and Patrick Laidlaw succeeded in extracting influenza viruses from humans. The only way to keep the virus in culture was to infect ferrets with throat swabs from humans and then transfer the infection from ferret to ferret before the infected animal became immune. Later they switched to mice.
The development of a vaccine
In 1935, Wilson Smith showed that the virus reproduced well in fertilized chicken eggs , a technique that had been used with other viruses since 1932. This means that research was no longer solely dependent on laboratory animals. Joseph Stokes of the University of Pennsylvania then began the first animal experiments with influenza vaccines between 1936 and 1938 . The results were unclear. The antibiotics developed in the 1940s prevented the disruptive growth of bacteria in the inoculated hen's eggs. In addition, various techniques have been worked out to enrich the viruses.
During World War II , a vaccine that looks modern was developed for the United States Department of Defense . It contained three different virus strains, was concentrated and inactivated with formalin . The following vaccine test was also innovative: The subjects who were to be vaccinated were selected at random ( randomized ), the control group received a placebo , and the study was double-blind . The vaccine produced excellent results in the 1943 flu wave, and by 1945 all US soldiers were vaccinated. In a major influenza B virus epidemic earlier this year, the vaccine was also shown to be highly effective. In 1945, various manufacturers in the US were also licensed to produce influenza vaccines for civilians.
More outbreaks
The 1947 and 1951 epidemics
In 1947 there was a recombination of gene segments within the subtype H1N1, which led to a complete failure of the vaccine. In particular, the hemagglutinin differed in numerous amino acids from its predecessor (reference strain: A / Fort Monmouth (FM) / 1/1947, at that time mostly called FM-1 virus). Although the virus spread around the world, mortality remained relatively low. In the long term, this event led to the realization that it would be impossible to produce an influenza vaccine that would protect in the long term. The vaccination has to be adapted to the circulating influenza strains every year , mainly because of the gradual change in genes through point mutations ( genetic drift ). The World Health Organization , founded in 1948, set up a monitoring system for this.
Another major epidemic occurred in 1950-51, with mortality rates in the UK and Canada even exceeding the 1957 and 1968 pandemics . In 1957, the H1N1 virus disappeared from human existence and was replaced by the Asian flu virus H2N2 . It has therefore been concluded that H1 and H2 viruses cannot coexist in a population.
The classic swine influenza is spreading
Classical swine influenza must also have spread throughout the world during or after World War II . It was found in Canada and Brazil, all of Asia, Kenya, Great Britain, Czechoslovakia and in 1959 also in the Federal Republic of Germany. It then disappeared from Europe and did not reappear until 1976 in an outbreak in northern Italy. The viruses isolated there turned out to be closely related to North American strains, so that the disease was probably imported from the USA. It remained limited to northern Italy until 1979 and then spread across Western Europe, Scandinavia and Great Britain. In the United States and Asia, too, classic swine influenza remained the dominant form of influenza in pigs until the 1990s.
The H1N1 viruses circulating among humans can sporadically also infect pigs, as documented in Budapest in 1941, but then do not establish themselves among pigs. The exception is the Spanish flu pathogen, which led to classic swine influenza.
In humans, H1N1 viruses are subject to constant change , caused by point mutations and promoted by the selection pressure of the immune system. The virus of classic swine influenza, on the other hand, has remained genetically stable for decades. The mutations are not uncommon, but the selection always works undisturbed in the direction of optimal adaptation to the host, since in pig farming young, not yet immune pigs are constantly introduced into the herd. One researcher observed that influenza viruses circulated within a pig herd for an entire year.
The "swine flu" of 1976

In the following, “swine flu”, in contrast to swine influenza, describes a disease in humans. In 1976 there was a local outbreak of variant A / New Jersey / 1/1976 among US soldiers in Fort Dix , New Jersey .
The United States health authorities assessed the risk of recurrence of a disease the size of the Spanish flu to be so great that they launched a vaccine production program to protect every citizen of the United States. The first vaccinations took place on October 1, 1976, and by mid-December, 40 million Americans had been vaccinated - the largest vaccination campaign in history by then. In Europe, only the Netherlands joined the vaccination program. At the same time it became known that some people who had been vaccinated had developed Guillain-Barré syndrome ; to this day it is controversial whether this is actually a side effect of the vaccination. There was a spate of claims for damages totaling $ 3.5 billion.
It is likely that pigs were the source of the influenza virus in Fort Dix, but this has never been proven.
The 1977 Russian flu
In May 1977, an H1N1 virus outbreak occurred in Tianjin , China, after the subtype had disappeared for twenty years. By November the epidemic had spread to the Soviet Union - which is where it got its name - and to Hong Kong. The Russian flu virus was found to be nearly identical to the viruses circulating prior to 1957, specifically A / Roma / 1949, so it is widely believed that it was accidentally released from a laboratory. However, influenza viruses remain genetically stable in birds so that the Russian flu virus could have survived in this reservoir between 1957 and 1977.
H1N1 viruses have been circulating in mankind since 1977 along with the H3N2 viruses from the 1968 Hong Kong flu . H1 and H3 viruses can therefore coexist with each other. The mortality in an influenza season in which H1N1 dominates is rather low. In addition, the long-term trend in mortality associated with influenza is falling in Germany.
H1N1 viruses in humans change only slowly due to genetic drift, which is shown by the fact that for them the composition of the vaccine only rarely needs to be changed. One of the three subtypes in the vaccine is traditionally reserved for an H1N1 strain, with A / Brisbane / 59/2007 scheduled for the 2009/2010 season in the northern hemisphere.
2009 H1N1 pandemic
In the spring of 2009, a new subtype " A / California / 7/2009 (H1N1) " of the H1N1 virus first spread in North America and soon afterwards caused a pandemic . According to a study published in 2016, the pandemic variant of the virus (pdmH1N1) originated in pigs in Mexico.
The virus itself, but also the vaccination, can in rare cases lead to narcolepsy . According to the EudraVigilance database of the European Medicines Agency, more than 1,300 cases of vaccinated people were known between 2010 and January 2015 , including some from Germany. As of November 28, 2016, 86 people in Germany who had been vaccinated with Pandemrix had been reported to the Paul Ehrlich Institute as "suspected narcolepsy cases". In Sweden, a law on compensation for victims was passed in 2016.
A general nomenclature for influenza viruses
In order to express the knowledge that influenza viruses can be transmitted across species boundaries, the World Health Organization implemented a nomenclature from 1980 based on the two most important surface antigens, hemagglutinin (= H) and neuraminidase (= N). The type of host is no longer taken into account.
The reconstruction of the causative agent of the Spanish flu
A working group led by Jeffery Taubenberger , at that time at the Armed Forces Institute of Pathology in Washington, DC, began sequencing the genome of the Spanish flu pathogen using tissue samples from three different sources . This enabled them to confirm that it was actually an H1N1 virus. A virus that has been experimented with since then could also be reconstructed from the genetic sequence. It has been shown to be fatal to mice, ferrets, and macaques, while it only causes mild disease in pigs.
In animal experiments on macaques, the reconstructed pathogen triggered an overreaction in the innate branch of the immune system, which reacts to viral infections. Your body produced so many cytokines and chemokines ("cytokine storm") that the regulation of the immune defense collapsed. The same sequence is likely for the human victims of the Spanish flu and would explain why especially young adults with well developed immune systems died.
Influenza A virus 1918 (H1N1) was assigned to risk group 3 in 2006 , while it was previously classified in risk group 2. Working with it may therefore only be carried out under the strict safety precautions of protection level 3 . The assignment only applies to the Spanish flu virus, the influenza A virus H1N1 derived therefrom, such as the classic swine influenza virus H1N1, the Eurasian swine influenza virus H1N1 or the H1N1 virus involved in the 2009/10 pandemic, remain due to the Biological Agents Ordinance in conjunction with the TRBA ( Technical rules for biological agents) 466 assigned to risk group 2.
H1N1 viruses that are derived directly from pathogens in birds
All of the H1N1 viruses discussed so far form a community of descent that goes back to the Spanish flu pathogen. However, since 1979, H1N1 viruses, which have been transmitted directly from birds (English: avian-like swine influenza ) have dominated European pigs . They have been shown to be closely related to duck H1N1 viruses. Since all gene segments are from birds, it appears that a complete virus has been transmitted. In Europe, these viruses, which originate from the bird kingdom, have replaced the classic swine influenza within a few years. Independently of this, this process appears to have taken place a second time in Southeast Asia in 1993; there the viruses originating from birds still circulate together with the classic swine influenza. The original avian viruses from Europe and Southeast Asia have proven to be related to each other.
Influenza virus transmission from pigs to birds
H1N1 influenza virus transmission from pigs to turkeys has been reported in North America. Multiple diseased turkeys immediately after the outbreak of influenza in pigs, and genetic studies have shown a lively exchange of influenza A virus between the two species. In one case, a human was also infected by a swine influenza virus through a turkey. In Europe, the H1N1 swine influenza viruses, originally derived from ducks, have also infected turkeys. So there is a possibility that pigs, humans, turkeys and ducks will infect each other over and over again.
H1N1 viruses as recombinants from various sources
The pig as a potential "mixing vessel"
Pigs are the only mammalian species that are kept in large numbers and in which both avian and human influenza viruses can reproduce. This is because the trachea of pigs is lined with cells that carry two types of sialic acid , which can be used as a docking point for influenza viruses in humans or birds. Since the genome of the influenza A virus is distributed over eight segments, these segments can reassort themselves in the event of a double infection with different viruses . Pigs form a potential "mixing vessel" ( mixing vessel ) for different variants of viruses and so a potential link between birds and humans.
Transmission of swine influenza virus from pigs to humans
The role pigs played in the development of the Spanish flu is controversial. Since the 1970s, more than 50 human infections with classic swine influenza have been documented in the scientific literature. It is known from studies on serum from occupationally exposed people that a much larger number can be expected.
Triple combinants
Until the 1990s, the classic swine influenza virus was the most common influenza virus among North American pigs. In the late 1990s, it was replaced by various strains and subtypes (H1N1, H3N2 and H1N2) that contained gene segments from three different sources (English: triple-reassortant swine influenza ). The basis was usually the H1 swine influenza virus of North American origin with other gene segments from influenza viruses from birds and humans.
In December 2005, infection of a human by such a triple combination was observed for the first time in Wisconsin . In June 2007, the United States declared such infections notifiable. By February 2009, a total of eleven cases were known from the Midwest and the south of the USA (ten of them H1N1, one H1N2). Nine of those affected had been near pigs. The patients were very young, the median age was 10 years.
A triple combination with “pandemic potential” was detected between 2011 and 2018 and since 2016 predominantly in pig herds in the People's Republic of China. The genes of the H1N1 variant called "G4 EA H1N1" come partly from a line known from European and Asian birds, partly from the pandemic H1N1 virus from 2009 (“swine flu”) and partly from a line known from North America, which in turn comes from is composed of several origins. According to a study published in June 2020 in the journal PNAS , more than 10 percent of pig farmers (35 of 338 people examined) are already infected. It was also pointed out that infection with seasonal influenza does not protect against “G4 EA H1N1”.
Quadruple combinants
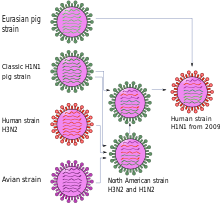
The first and so far only quadruple combination was discovered in 2009. Although it was already adapted to humans, see pandemic influenza 2009 , it was often referred to as "swine flu" because it originated from swine influenza viruses and probably in pigs.
The first scientific publication describes the new virus variant as a genetic new combination of two virus lines of swine influenza , one North American and one Eurasian. Three of the eight gene segments - NP, NS and one coding for H1 hemagglutinin - go back to the classic swine influenza virus of 1918. Two gene segments - PB2 and PA - passed from birds to North American pigs around 1998 and PB1 from birds to humans around 1968 and from there to pigs around 1998. This combination of six gene segments is found in triple combinants that are common in North America. In contrast, the two remaining gene segments that encode an N1 neuraminidase and the matrix proteins M1 and M2 are derived from the variant originally distributed among birds , which has been distributed among Eurasian pigs since 1977 but was never detected in America. The gene for the matrix protein contains a mutation that makes the virus resistant to the influenza drugs amantadine and rimantadine .
In Canada, pigs were again infected with the new variant by humans. The symptoms are similar to those of classic swine influenza. In the event of an outbreak in Argentina, it is not yet clear whether it is the classic swine influenza or the new H1N1 variant.
See also
literature
- Report on the Pandemic of Influenza, 1918-1919 . Ministry of Health (Ed.). In: Reports on Public Health and Medical Subjects . Volume 4, His Majesty's Stationery Office, London 1920 (best contemporary survey of the Spanish flu)
- Ian H. Brown: The epidemiology and evolution of influenza viruses in pigs . In: Veterinary Microbiology . Volume 74, No. 1–2, 2000, pp. 29–46, PMID 10799776 (review article on swine influenza)
Web links
- Viruses razor-ears in the microcosm. Planet Wissen - broadcast on January 14, 2011
Individual evidence
- ↑ ICTV Master Species List 2018b.v2 . MSL # 34, March 2019
- ↑ a b ICTV: ICTV Taxonomy history: Akabane orthobunyavirus , EC 51, Berlin, Germany, July 2019; Email ratification March 2020 (MSL # 35)
- ↑ Negative Sense RNA Viruses: Orthomyxoviridae , in: ICTV 9th Report (2011)
- ↑ Edwin O. Jordan: Epidemic Influenza. A survey . American Medical Association, Chicago 1927, p. 229.
- ^ ID Mills: The 1918-1919 Influenza Pandemic. The Indian Experience . In: The Indian Economic and Social History Review . Volume 23, No. 1, 1986, pp. 1-40.
- ↑ Jeffery K. Taubenberger et al. : Characterization of the 1918 influenza virus polymerase genes . In: Nature , Volume 437, 2005, pp. 889-893.
- ^ Mark J. Gibbs and Adrian J. Gibbs: Was the 1918 pandemic caused by a bird flu? as well as Janis Antonovics , Michael E. Hood and Christi Howell Baker: Was the 1918 flu avian in origin? both in: Nature . Volume 440, 2006, pp. E8f.
- ^ JWH Chun: Influenza Including its Infection Among Pigs . In: The National Medical Journal of China . Volume 5, No. 1, 1919, pp. 34-44.
- ^ JS Koen: A Practical Method for Field Diagnosis of Swine Diseases . In: American Journal of Veterinary Medicine . Volume 14, 1919, pp. 468-470.
- ↑ http://www.pandemicinfluenza.org/about.html ( Memento from January 30, 2010 in the Internet Archive ) Katzelab
- ↑ E. Eckardt, H. Flohn and HJ Jusatz: Spread and course of the flu epidemic 1933 as a function of meteorological and geographical factors. In: Journal of Hygiene and Infectious Diseases. Volume 118, 1936, pp. 64-91, doi: 10.1007 / BF02177495 .
- ^ Richard E. Shope: Swine Influenza. III. Filtration Experiments and Etiology . In: Journal of Experimental Medicine . Volume 54, 1931, pp. 373-385.
- ^ Wilson Smith, CH Andrewes and PP Laidlaw: A Virus Obtained From Influenza Patients . In: The Lancet . Volume 222, 1933, pp. 66-68.
- ^ WIB Beveridge: Influenza. The Last Great Plague . Heinemann, London 1977, p. 9.
- ^ John M. Eyler: De Kruif's Boast: Vaccine Trials and the Construction of a Virus . In: Bulletin of the History of Medicine . Volume 80, No. 3, 2006, pp. 409-438.
- ↑ Martha I. Nelson et al. : Multiple Reassortment Events in the Evolutionary History of H1N1 Influenza A Virus Since 1918 . In: PLoS Pathogens . Volume 4, No. 2, 2008.
- ↑ Miklós Dreguss: About the infection of pigs with human influenza virus . In: Archives of Virology . Volume 3, No. 1-4, 1944, pp. 35-48.
- ↑ JC Gaydos et al. : Swine influenza A outbreak, Fort Dix, New Jersey, 1976. In: Emerging Infectious Diseases . Volume 12, 2006, pp. 23-28; Full text (in English)
- ↑ Gina Kolata: Influenza. The hunt for the virus. S. Fischer, Frankfurt 2001, pp. 153-213
- ↑ Martha I. Nelson et al. : Multiple Reassortment Events in the Evolutionary History of H1N1 Influenza A Virus Since 1918 . In: PLoS Pathogens . Vol. 4, No. 2, February 29, 2008, e1000012, doi: 10.1371 / journal.ppat.1000012 .
- ↑ Alan P. Kendal et al. : Antigenic Similarity of Influenza A (H1N1) Viruses from Epidemics in 1977–1978 to 'Scandinavian' Strains Isolated in Epidemics of 1950–1951 . In: Virology . Volume 89, 1978, pp. 632-636.
- ↑ Influenza-associated mortality in Germany 1985–2006 . In: Epidemiological Bulletin . No. 35, 2007, pp. 325-327.
- ↑ Ignacio Mena et al. : Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. In: eLife. Online publication from June 28, 2016, doi: 10.7554 / eLife.16777
-
↑ Syed Sohail Ahmed et al .: Antibodies to influenza nucleoprotein cross-react with human hypocretin receptor 2. In: Science Translational Medicine. Volume 7, No. 294, 2015, p. 294ra105, doi: 10.1126 / scitranslmed.aab2354
How H1N1 can trigger sleeping sickness . Zeit Online , July 1, 2015 - Jump up ↑ Influenza vaccination: How Pandemrix causes narcolepsy. On: aerzteblatt.de of July 2, 2015
- ↑ Current information on narcolepsy in relation to A / H1N1 influenza vaccination. On: pei.de from November 28, 2016
- ↑ Damage after swine flu vaccination: Sweden compensates narcolepsy patients . Spiegel Online , May 13, 2016
- ↑ Darwyn Kobasa: Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus . In: Nature . Volume 445, 2007, pp. 319-323.
- ↑ TRBA (Technical Rules for Biological Agents) 462: Classification of viruses in risk groups. In: Website of the Federal Institute for Occupational Safety and Health (BAuA) . April 25, 2012, accessed May 13, 2014 .
- ^ Hermann Müller: The classic avian influenza ("highly pathogenic avian influenza"; "bird flu") - background information on the current epidemic in Southeast Asia and references to information on the Internet. (PDF) Director of the Institute for Virology, Faculty of Veterinary Medicine, University of Leipzig, status: 2004
- ↑ Vivek Shinde et al. : Triple-Reassortant Swine Influenza A (H1) in Humans in the United States, 2005-2009. In: The New England Journal of Medicine . Volume 360, No. 25, 2009, pp. 2616-2625, doi: 10.1056 / NEJMoa0903812 .
- ↑ New type of swine flu with pandemic potential. On: science.orf.at from June 30, 2020.
- ↑ Honglei Sun et al .: Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. In: PNAS. Online advance publication of June 29, 2020, doi: 10.1073 / pnas.1921186117 .
- ↑ Swine flu strain with human pandemic potential increasingly found in pigs in China. On: sciencemag.org from June 29, 2020.
- ^ RJ Garten, CT Davis et al. : Antigenic and Genetic Characteristics of Swine-Origin 2009 A (H1N1) Influenza Viruses Circulating in Humans. In: Science . July 10, 2009, Volume 325, No. 5937, pp. 197-201, doi: 10.1126 / science.1176225 .
- ↑ Case report of the Canadian outbreak on the World Organization for Animal Health OIE website
- ^ Case report of the Argentine outbreak on the World Organization for Animal Health OIE website