SDS-PAGE
SDS-PAGE (abbreviation for English s odium d odecyl s ulfate p oly a crylamide g el e lectrophoresis , sodium dodecyl sulfate-polyacrylamide gel electrophoresis ) is a variant of the polyacrylamide gel electrophoresis , an analytical method of biochemistry for separation of substance mixtures according to the molecular mass in an electric field .
This discontinuous electrophoretic system developed by Ulrich K. Laemmli enables a good separation of proteins with molecular weights between 5 and 250 kDa . The publication in which it was described is the most cited paper by a single author and the second most cited overall.
properties
The SDS-PAGE is used to analyze proteins . A discontinuous gel based on polyacrylamide is used as the separation medium (also known as matrix) in this type of electrophoresis . SDS ( sodium dodecyl sulfate ) is also used. This anionic surfactant ( detergent ) covers the intrinsic charges of proteins. Per gram of protein, around 1.4 grams of SDS bind constantly, corresponding to one SDS molecule per two amino acids , so that the proteins have a constant negative charge distribution. The intrinsic charges of the proteins are negligible under the SDS load, the positive charges are also greatly reduced in the basic pH range of a separating gel according to Laemmli. The negative charges of the SDS cause their mutual repulsion, which together with the denaturation caused by boiling leads to a linearization of the previously folded proteins. This allows a separation according to the chain length, proportional to the molecular mass, because longer proteins are more strongly retained in the gel than shorter ones.
Above a certain concentration, SDS tends to form spherical micelles in aqueous solutions . SDS occurs in solutions from the critical micelle concentration of 7 to 10 millimolar simultaneously as single molecules ( monomer ) and as micelles, below that SDS occurs only individually. At the critical micelle concentration, a micelle consists of around 62 SDS molecules. However, only SDS monomers bind to proteins via hydrophobic interactions , while the SDS micelles that are anionic on the outside do not take up any protein. In SDS concentrations above 0.1 millimolar, the unfolding of proteins begins, above 1 mM most proteins are denatured . Because of the strong denaturing effect of SDS and the subsequent breakdown of protein complexes , it is usually not possible to determine quaternary structures with SDS . Exceptions are e.g. B. proteins previously stabilized by covalent cross-linking and the SDS-resistant protein complexes, which are also stable in the presence of SDS (the latter, however, only at room temperature). SDS resistance is based on metastability : In order to denature the resistant complexes, a high activation energy is required, which is achieved by heating. Although the native, fully folded, SDS-resistant protein does not have sufficient stability under the conditions, the chemical equilibrium of denaturation is only established slowly at room temperature. In addition to resistance to SDS, stable protein complexes are also characterized by stability against proteases and an increased biological half-life .
Alternatively, a polyacrylamide gel electrophoresis with the cationic surfactants CTAB in a CTAB-PAGE or 16-BAC in a BAC-PAGE can also be used in some cases.
Physical basics
In an electric field, charged particles in solution migrate to the pole with the opposite charge . Due to the loading of proteins with SDS, they are afflicted with several negative charges, which is why proteins migrate to the plus pole in SDS-PAGE. The use of an uncharged polymeric hydrogel (polyacrylamide) also increases the sieving effect. The pore size in the hydrogel is determined by the concentration of monomer and crosslinker .
Procedure
The SDS-PAGE method consists of gel production, sample preparation, electrophoresis, protein staining or a Western blot and analysis of the band pattern generated.
Gel production

If different buffers are used in the gel (discontinuous gel electrophoresis), the gels are prepared up to one day before the electrophoresis so that the diffusion does not lead to a mixing of the buffers. The gel is created by radical polymerization in a mold that consists of two sealed glass plates with spacers between the glass plates. The spacers have a thickness of 0.75 mm or 1.5 mm, which determines the loading capacity of the gel. For the pouring of the gel solution, the plates are usually clamped into a stand that temporarily seals the otherwise open underside of the glass plates with the two spacers. For the gel solution, acrylamide is mixed as a gel former (mostly 4% V / V in the stacking gel and 10–12% in the separating gel), methylenebisacrylamide as a crosslinker , collecting or separating gel buffer, and water and SDS. The polymerization is started by adding the catalyst TEMED and the radical initiator ammonium persulfate (APS). The solution is then poured between the glass plates without bubbles. Depending on the amount of catalyst and free radical initiator as well as the temperature, the polymerization takes between a quarter of an hour and several hours. The lower gel (separating gel) is first poured and covered with a few drops of a slightly water-soluble alcohol (mostly buffer-saturated butanol or isopropanol ), whereby the meniscus becomes bubble-free and is protected from the radical scavenger oxygen. After the separation gel has polymerized, the alcohol is tipped off and the remains are removed with filter paper. After adding APS and TEMED to the collecting gel solution, the collecting gel solution is poured onto the solid separating gel and a suitable sample comb is inserted without bubbles. After polymerisation, the sample comb is carefully pulled out, creating pockets for sample application. For a later use of the proteins for protein sequencing , the gels are often prepared on the day before electrophoresis in order to reduce reactions of unpolymerized acrylamide with cysteines in proteins.
Using a gradient mixer , gradient gels with a gradient of acrylamide (usually from 4 to 12%) can be poured, which have a larger separation range of the molar masses. Commercial gel systems (so-called pre-cast gels) mostly use the buffer substance BisTris with a pH value between 6.4 and 7.2 in both the collecting gel and the separating gel. These gels are delivered already poured and are ready for use immediately. Since they only use one buffer (continuous gel electrophoresis) and have an almost neutral pH value, they can be stored for several weeks. The more neutral pH slows down the hydrolysis and thus the breakdown of the polyacrylamide. Furthermore, there are fewer acrylamide-modified cysteines in the proteins. Due to the constant pH in the collecting and separating gel, there is no stacking effect. Proteins in BisTris gels cannot be stained with ruthenium complexes . This gel system has a comparatively large separation area, which can be varied by using MES or MOPS in the running buffer.
Sample preparation
During sample preparation, sample buffer and thus excess SDS is added to the proteins and the sample is then heated to 95 ° C for five minutes in order to break up secondary and tertiary structures by breaking hydrogen bonds and stretching the molecules. Optionally, disulfide bridges can be cleaved by reduction. For this purpose, reducing thiols such as β-mercaptoethanol (β-ME, 5% by volume), dithiothreitol (DTT, 10 millimolar) or dithioerythritol (DTE, 10 millimolar) are added to the sample buffer. Each sample is pipetted into its own pocket in the gel that has previously been submerged in electrophoresis buffer in the electrophoresis apparatus.
In addition to the samples, a size marker is usually loaded onto the gel. This consists of proteins of known size and thus enables the size of the proteins in the actual samples to be estimated with an accuracy of ± 10%, which migrate in parallel in different tracks of the gel. The size marker is often pipetted into the first or last pocket of a gel.
Electrophoresis

For separation, the denatured samples are loaded onto a gel made of polyacrylamide, which is placed in an electrophoresis buffer with suitable electrolytes . Then an electrical voltage (usually around 100 volts , 10–20 V per cm of gel length) is applied, which causes the negatively charged molecules to migrate through the gel in the direction of the anode (positive pole). The gel acts like a sieve. Small proteins migrate relatively easily through the mesh of the gel, while large proteins tend to be retained and therefore migrate more slowly through the gel. The electrophoresis takes between three quarters of an hour and several hours, depending on the voltage used and the length of the gel.
The fastest migrating proteins (with a molar mass below 5 KDa) form the mobile phase front with the anionic components of the electrophoresis buffer that also migrate through the gel. The area of the solvent front is made visible by adding the comparatively small, anionic dye bromophenol blue to the sample buffer. Because of the relatively small molecular size of bromophenol blue, it migrates faster than proteins. With the optical control based on the moving colored band, the electrophoresis can be ended before the dye and thus also the samples have completely migrated through the gel and leave it again.
The most frequently used method is discontinuous SDS-PAGE. In this process, the proteins first migrate into a collecting gel with neutral pH, in which they are concentrated, and then into a separating gel with basic pH, in which the actual separation takes place. Collecting gel and separating gel differ in their pore size (4-6% T and 10-20% T), ionic strength and pH values (pH 6.8 and pH 8.8). An SDS-containing TRIS - glycine - chloride - buffer system is often used as the electrolyte . Glycine forms predominantly the zwitterionic form at a neutral pH value ; at high pH values the glycinations lose positive charges and become predominantly anionic. In stacking the smaller negatively charged chloride ions migrate in front of the proteins (engl. Leading ions , leading ions ') and the slightly larger, negative and partially positively charged glycinate ion by (engl. Trailing ion , the following ions'), while in the comparatively basic separation gel, both ions migrate in front of the proteins. The pH gradient between the collecting gel and the separating gel buffer leads to a stacking effect at the boundary between the collecting gel and the separating gel, as the glycinate partially loses its retarding positive charge as the pH value rises and then, as the previous ion, overtakes the proteins while migrating and becomes the leading ion which makes the bands of the various proteins visible after staining become narrower and sharper ( stacking effect ). The TRIS tricine buffer system from Schägger and von Jagow is suitable for separating smaller proteins and peptides due to the higher spread of the proteins in the range from 0.5 to 50 KDa .
coloring
At the end of the electrophoretic separation, all proteins are sorted according to size and can then be used by further processes ( protein staining such as Coomassie staining ( easy to carry out, most frequently used), silver staining (highest sensitivity), stains-all staining, the Amido black 10 B staining, the Fast Green FCF staining, the fluorescent staining such as the Epicocconon staining and the SYPRO orange staining as well as immunological evidence such as e.g. Western blot) can be made visible. The fluorescent stains have a comparatively higher linearity between the amount of protein and the color intensity of about three powers of ten above the detection limit , ie the amount of protein can be estimated from the color intensity. If the fluorescent protein dye trichloroethanol is used, there is no subsequent protein staining if it was added to the gel solution and the gel was irradiated with UV light after electrophoresis .
analysis
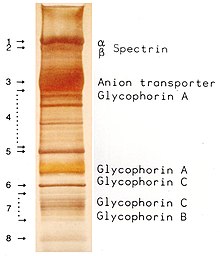
A protein staining gives the gel with the separated proteins a documentable band pattern of the various proteins. Glycoproteins adsorb SDS to the glycosylations more unevenly, which results in broader and fuzzier bands. Membrane proteins because of their transmembrane domain often built up from the more hydrophobic amino acids, have a lower solubility in aqueous solutions tend to bind lipids and tend due to hydrophobic effects to precipitation in aqueous solutions, unless sufficient detergent present. In the case of membrane proteins, this precipitation manifests itself in the SDS-PAGE as a “tail formation” above the band of the transmembrane protein, on the other hand more SDS can be used (by using more or more concentrated sample buffer) and the amount of protein reduced during the gel application. If the gel is overloaded with a soluble protein, this manifests itself in a semicircular band of this protein (e.g. in the marker lane in the figure at 66 KDa), whereby other proteins with similar molar masses can be covered. A low contrast (as in the marker lane in the figure) between the bands within a lane indicates either the presence of many proteins with intervening molecular weights (low purity ) or, if in the case of purified proteins there is a lack of contrast only below a band, a proteolytic one Degradation of the protein, which first manifests itself in degradation bands and after further degradation also in a homogeneous color (“smear”) below a band. The documentation of the band pattern is mostly done by taking photos or scanning. A gel extraction can be carried out for the subsequent recovery of the molecules in individual bands .
Archiving
After protein staining and documentation of the band pattern, the polyacrylamide gel can be dried for archiving. Proteins can be extracted from it at a later time. The gel is either placed in a drying frame (with or without heat supply) or in a vacuum gel dryer. The drying frame consists of two parts, one of which serves as a base for a wet cellophane film on which the gel and a one percent glycerol solution are placed. A second wet cellophane sheet is placed on top of it without bubbles, the second frame part is put on and the frame is sealed with a couple of clamps. The removal of the air bubbles avoids fragmentation of the gel during drying. The water contained evaporates through the cellophane film. A vacuum gel dryer, on the other hand, creates a negative pressure and heats the gel to around 50 ° C.
Molecular weight determination
With a more precise determination of the molar mass , the relative widths of the individual protein bands in the separating gel are measured. The measurements are usually carried out in triplicates to increase the accuracy. The relative tracking (also Rf value , Rm value ) is the quotient of the tracking of the band of the protein in question and the tracking of the protein front. The widths of the bands and the protein front are measured from the beginning of the separating gel. The distance of the protein front corresponds approximately to the distance of the bromophenol blue contained in the sample buffer . The relative distances of the proteins of the size marker are plotted semi-logarithmically against their known molar masses. The molar mass of an unknown protein can be determined on the basis of its relative distance by comparison with the linear range of the graph generated or by calculation using a regression analysis . Bands of proteins with glycosylations may be more blurred. Proteins with many basic amino acids (e.g. histones ) can lead to an overestimation of the molar mass or not migrate into the gel at all because they migrate more slowly or in the opposite direction during electrophoresis due to the positive charges. Correspondingly, many acidic amino acids can lead to an accelerated migration of a protein and an underestimation of the molar mass.
Applications
SDS-PAGE in combination with protein staining is used in biochemistry for the rapid and accurate separation and subsequent analysis of proteins. It has comparatively low equipment and reagent costs and is a comparatively simple method. Due to its low scalability , it is mainly used for analytical purposes and less for preparative purposes, especially when larger amounts of a protein are to be isolated.
In addition, SDS-PAGE is used in combination with Western Blot to determine the presence of a specific protein in a mixture of proteins - or for the analysis of post-translational modifications . Post-translational modifications of proteins can lead to a different relative mobility (ie a band shift ) or to a change in the binding of a detection antibody that is used in the Western blot (ie a band disappears or appears).
In protein mass spectrometry , SDS-PAGE is a widely used method for sample preparation prior to spectrometry, mostly with an in-gel digestion . With regard to the determination of the molecular mass of a protein, SDS-PAGE is somewhat more accurate than analytical ultracentrifugation , but less accurate than mass spectrometry or - without taking post-translational modifications into account - a calculation of the protein molecular mass from the DNA sequence .
In medical diagnostics, SDS-PAGE is used, among other things, as part of an HIV test and to investigate proteinuria . In the HIV test, HIV proteins are separated by SDS-PAGE and then detected by Western blot with HIV-specific antibodies of the patient, if they are present in his blood serum . An SDS-PAGE for proteinuria assesses the levels of various serum proteins in the urine, e.g. B. albumin , alpha-2 macroglobulin and IgG .
variants
SDS-PAGE is the most widely used method for the gel electrophoretic separation of proteins. The 2D gel electrophoresis sequentially combines isoelectric focusing or BAC-PAGE with SDS-PAGE. The native-PAGE is used when the native protein folding is to be maintained. As an alternative to SDS-PAGE, a BAC-PAGE or a CTAB-PAGE can be used to separate membrane proteins . For the electrophoretic separation of large protein complexes a can agarose gel electrophoresis are used, z. B. the SDD-AGE . Enzymes can partly be detected via their enzyme activity by zymography .
Alternatives
While SDS-PAGE is one of the more precise and inexpensive methods for protein separation and analysis, it denatures proteins. Where non-denaturing conditions are required, proteins are examined, for example, by native PAGE or various chromatographic methods with subsequent photometric quantification, such as affinity chromatography (or even tandem affinity purification ), size exclusion chromatography or ion exchange chromatography . Proteins can also be separated according to size in a tangential flow filtration or an ultrafiltration . Individual proteins can be isolated from a mixture by affinity chromatography or by a pull-down assay . Some historically early and cost-effective but messy separation methods , which are usually based on a series of extractions and precipitations and use cosmotropic molecules, are, for example, ammonium sulfate precipitation and polyethylene glycol precipitation . Because of their low cost, they are still used today to purify large quantities of proteins.
history
For the discovery of the principle of electrophoresis as the migration of charged and dissolved atoms or molecules in an electric field , Arne Tiselius was awarded the Nobel Prize in Chemistry in 1948 . The use of a solid matrix (initially paper disks) in zone electrophoresis improved the separation. The discontinuous electrophoresis from 1964 by L. Ornstein and BJ Davis made it possible to improve the separation through the stacking effect. The use of cross-linked polyacrylamide - hydrogels offered in contrast to the previously used paper discs or starch gels a higher stability of the gel and no microbial decomposition . The denaturing effect of SDS in continuous polyacrylamide gels and the resulting improvement in separation was first described in 1965 by David F. Summers in the working group of James E. Darnell for the separation of the proteins of the poliovirus . Today's variant of SDS-PAGE was described by Ulrich K. Laemmli in 1970 and used for the first time to characterize the proteins in the head of the bacteriophage T4 .
literature
- Hubert Rehm , Thomas Letzel: The Experimenter: Protein Biochemistry / Proteomics . 6th edition, Spektrum Akademischer Verlag, Heidelberg 2009, ISBN 978-3-8274-2312-2 .
Web links
- OpenWetWare: Protocol for BisTris SDS-PAGE
Individual evidence
- ↑ a b U. K. Laemmli: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. In: Nature . Vol. 227, 1970, pp. 680-685, doi : 10.1038 / 227680a0 , PMID 5432063 .
- ↑ Interview with Ulrich Laemmli . In: NZZ Folio . No. 11, 2005. Retrieved March 4, 2012.
- ↑ BJ Smith: SDS Polyacrylamide Gel Electrophoresis of Proteins. In: Methods in molecular biology (Clifton, NJ). Volume 1, 1984, pp. 41-55, doi : 10.1385 / 0-89603-062-8: 41 , PMID 20512673 .
- ^ R. Pitt-Rivers, FS Impiombato: The binding of sodium dodecyl sulphate to various proteins. In: The Biochemical journal. Volume 109, Number 5, October 1968, pp. 825-830, PMID 4177067 , PMC 1187034 (free full text).
- ↑ a b c J. A. Reynolds, Charles Tanford : Binding of dodecyl sulfate to proteins at high binding ratios. Possible implications for the state of proteins in biological membranes. In: Proceedings of the National Academy of Sciences . Volume 66, Number 3, July 1970, pp. 1002-1007, PMID 5269225 , PMC 283150 (free full text).
- ↑ Georgios Staikos, Anastasios Dondos: Study of the sodium dodecyl sulphate-protein complexes: evidence of their wormlike conformation by treating them as random coil polymers. In: Colloid and Polymer Science. 287, 2009, p. 1001, doi : 10.1007 / s00396-009-2059-3 .
- ↑ Nicholas J. Turro, Ahmad Yekta: Luminescent probes for detergent solutions. A simple procedure for determination of the mean aggregation number of micelles. In: Journal of the American Chemical Society. 100, 1978, p. 5951, doi : 10.1021 / ja00486a062 .
- ↑ Marta Manning, Wilfredo Colón: Structural basis of protein kinetic stability: resistance to sodium dodecyl sulfate suggests a central role for rigidity and a bias toward beta-sheet structure. In: Biochemistry . Vol. 43, 2004, pp. 11248-11254, PMID 15366934 .
- ↑ Engelbert Buxbaum: Cationic electrophoresis and electrotransfer of membrane glycoproteins. In: Analytical Biochemistry. Vol. 314, No. 1, 2003, pp. 70-76, PMID 12633604 , doi : 10.1016 / S0003-2697 (02) 00639-5 .
- ↑ Dianne T. Akin, Raymond Shapira, Joseph M. Kinkade Jr .: The determination of molecular weights of biologically active proteins by cetyltrimethylammonium bromide-polyacrylamide gel electrophoresis. In: Analytical Biochemistry. Vol. 145, No. 1, 1985, pp. 170-176, PMID 4003759 , doi : 10.1016 / 0003-2697 (85) 90343-4 .
- ^ RJ Simpson: CTAB-PAGE. In: Cold Spring Harbor protocols. Volume 2010, number 4, April 2010, p. Pdb.prot5412, doi : 10.1101 / pdb.prot5412 , PMID 20360366 .
- ↑ Joachim Hartinger, Katinka Stenius, Dagmar Högemann, Reinhard Jahn : 16-BAC / SDS-PAGE: a two-dimensional gel electrophoresis system suitable for the separation of integral membrane proteins. In: Analytical Biochemistry. Vol. 240, No. 1, 1996, pp. 126-133, PMID 8811889 , doi : 10.1006 / abio.1996.0339 .
- ^ J. Margolis, KG Kenrick: 2-dimensional resolution of plasma proteins by combination of polyacrylamide disc and gradient gel electrophoresis. In: Nature. Volume 221, Number 5185, March 1969, pp. 1056-1057, PMID 5774398 .
- ↑ a b c J. P. Hachmann, JW Amshey: Models of protein modification in Tris-glycine and neutral pH Bis-Tris gels during electrophoresis: effect of gel pH. In: Analytical biochemistry. Volume 342, number 2, July 2005, pp. 237-245, doi : 10.1016 / j.ab.2005.04.015 , PMID 15935323 .
- ↑ J. Wiltfang, N. Arold, V. Neuhoff: A new multiphasic buffer system for sodium dodecyl sulfate-polyacrylamide gel electrophoresis of proteins and peptides with molecular masses 100,000-1000, and their detection with picomolar sensitivity. In: Electrophoresis. Volume 12, Number 5, May 1991, pp. 352-366, doi : 10.1002 / elps.1150120507 , PMID 1718736 .
- ↑ A. Penna, M. Cahalan: Western blotting using the Invitrogen Novex NuPage Until Tris minigel. In: Journal of visualized experiments: JoVE. Number 7, 2007, p. 264, doi : 10.3791 / 264 , PMID 18989435 , PMC 2565856 (free full text).
- ↑ J. Moebius, K. Denker, A. Sickmann: Ruthenium (II) tris-bathophenanthroline disulfonate is well suitable for Tris-Glycine PAGE but not for Bis-Tris gels. In: Proteomics. Volume 7, Number 4, February 2007, pp. 524-527, doi : 10.1002 / pmic.200600642 , PMID 17309097 .
- ^ Ian M. Rosenberg: Protein Analysis and Purification: Benchtop Techniques . Springer Science & Business Media, December 22, 2006, ISBN 978-0-8176-4412-3 , p. 103.
- ↑ Hermann Schägger, Gebhard von Jagow: Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. In: Analytical Biochemistry . Vol. 166, 1987, pp. 368-379, doi : 10.1016 / 0003-2697 (87) 90587-2 , PMID 2449095 .
- ^ S. Fazekas de St. Groth, RG Webster, A. Datyner: Two new staining procedures for quantitative estimation of proteins on electrophoretic strips . In: Biochimica et Biophysica Acta . 71, 1963, pp. 377-391. doi : 10.1016 / 0006-3002 (63) 91092-8 . PMID 18421828 .
- ^ A b c Wilson CM: Studies and critique of Amido Black 10B, Coomassie Blue R, and Fast Green FCF as stains for proteins after polyacrylamide gel electrophoresis. . In: Anal Biochem . 96, No. 2, 1979, pp. 263-78. PMID 89822 .
- ↑ CR Merril, RC Switzer, ML Van Keuren: Trace polypeptides in cellular extracts and human body fluids detected by two-dimensional electrophoresis and a highly sensitive silver stain. In: Proc Natl Acad Sci USA . 76 (9), 1979, pp. 4335-4339. PMID 92027 .
- ↑ RC Switzer, CR Merril, S. Shifrin: A highly sensitive silver stain for detecting proteins and peptides in polyacrylamide gels . In: Anal Biochem . tape 98 , no. 1 , September 1979, p. 231-237 , doi : 10.1016 / 0003-2697 (79) 90732-2 , PMID 94518 .
- ↑ T. Rabilloud et al .: Improvement and simplification of low-background silver staining of proteins by using sodium dithionite. In: Electrophoresis. 9 (6), 1998, pp. 288-291. PMID 2466660 .
- ↑ T. Rabilloud: A comparison between low background silver diammine silver nitrate and protein stains. In: Electrophoresis. 13, 1992, pp. 429-439. PMID 1425556 .
- ↑ C. Lelong, M. Chevallet, S. Luche, T. Rabilloud: Silver staining of proteins in 2DE gels. In: Methods Mol Biol. 519, 2009, pp. 339-350. PMID 19381593 .
- ↑ H. Blum, H. Beier, HJ Gross: Improved silver staining of plant protein, RNA & DNA in PAA gels. In: Electrophoresis. 8, 1987, pp. 93-99.
- ↑ Christian P. Moritz, Sabrina X. Marz, Ralph Reiss, Thomas Schulenborg, Eckhard Friauf: Epicocconone staining: a powerful loading control for Western blots. In: Proteomics . PMID 24339236 .
- ↑ Aleksandr Petrovich Demchenko: Advanced Fluorescence Reporters in Chemistry and Biology III: Applications in Sensing and Imaging Volume 3 of Advanced Fluorescence Reporters in Chemistry and Biology . Springer 2011, ISBN 978-3-642-18035-4 , pp. 179ff.
- ↑ S. Gallagher, D. Chakavarti: staining proteins in gels. In: Journal of visualized experiments: JoVE. Number 17, 2008, ISSN 1940-087X , 760, doi : 10.3791 / 760 , PMID 19066521 . PMC 3253607 (free full text).
- ↑ CM Wilson: Staining of proteins on gels: comparisons of dyes and procedures. In: Methods in enzymology. Volume 91, 1983, ISSN 0076-6879 , pp. 236-247, PMID 6190068 .
- ↑ Carol L. Ladner, Jing Yang, Raymond J. Turner, Robert A. Edwards: Visible fluorescent detection of proteins in polyacrylamide gels without staining. In: Analytical Biochemistry Vol. 326, 2004, pp. 13-20, PMID 14769330 .
- ↑ Jennifer E. Gilda, Aldrin V. Gomes: Stain-Free total protein staining is a superior loading control to β-actin for Western blots. In: Analytical Biochemistry Vol. 440, 2013, pp. 186-188, PMID 23747530 .
- ^ Cryo-EM Part A: Sample Preparation and Data Collection . Academic Press, September 30, 2010, ISBN 978-0-08-095695-4 , pp. 28-.
- ^ Richard R Burgess, Murray P. Deutscher: Guide to Protein Purification . Academic Press, November 3, 2009, ISBN 978-0-08-092317-8 , p. 184.
- ^ Philip LR Bonner, Alan J. Hargreaves: Basic Bioscience Laboratory Techniques: A Pocket Guide . John Wiley & Sons, August 24, 2011, ISBN 978-1-119-95644-0 , p. 140.
- ^ Martin Holtzhauer: Basic Methods for the Biochemical Lab . Springer Science & Business Media, September 13, 2006, ISBN 978-3-540-32786-8 , p. 243.
- ↑ WJ van Venrooij, Ravinder N. Maini: Manual of Biological Markers of Disease . Springer Science & Business Media, 6 December 2012, ISBN 978-94-011-1670-1 , p. 50.
- ↑ Y. Guan, Q. Zhu, D. Huang, S. Zhao, L. Jan Lo, J. Peng: An equation to estimate the difference between theoretically predicted and SDS PAGE-displayed molecular weights for an acidic peptide. In: Scientific reports. Volume 5, 2015, p. 13370, doi : 10.1038 / srep13370 , PMID 26311515 , PMC 4550835 (free full text).
- ↑ Jan-Christer Janson: Protein Purification: Principles, High Resolution Methods, and Applications . John Wiley & Sons, January 3, 2012, ISBN 978-1-118-00219-3 .
- ↑ Mohamed A. Desai: Downstream Processing of Proteins: Methods and Protocols . Springer Science & Business Media, 2000, ISBN 978-1-59259-027-8 , p. 35.
- ↑ Ghosh Raja: Protein Bioseparation Using Ultrafiltration: Theory, Applications And New Developments . World Scientific, June 11, 2003, ISBN 978-1-78326-126-0 , p. 142.
- ↑ T. Pederson: Turning a PAGE: the overnight sensation of SDS-polyacrylamide gel electrophoresis. In: FASEB journal: official publication of the Federation of American Societies for Experimental Biology. Volume 22, Number 4, April 2008, pp. 949-953, doi : 10.1096 / fj.08-0402ufm , PMID 18378803 .
- ↑ L. Ornstein, BJ Davis: Disc Electrophoresis -1. Background and Theory. In: Ann NY Acad Sci. Volume 121, 1964, pp. 321-349.
- ↑ David F. Summers, Jacob V. Maizel, James E. Darnell: Evidence for virus-specific noncapsid proteins in poliovirus-infected HeLa cells. In: Proceedings of the National Academy of Sciences . Volume 54, Number 2, August 1965, pp. 505-513, PMID 4285933 , PMC 219696 (free full text).