Archaea
| Archaea | ||||
|---|---|---|---|---|

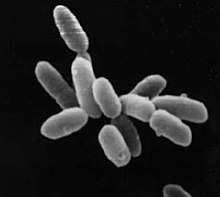
Archaeon of the genus Sulfolobus , infected with the Sulfolobus virus STSV1. Scale = 1 μm. |
||||
| Systematics | ||||
|
||||
| Scientific name | ||||
| Archaea | ||||
| Otto Kandler & Mark L. Wheelis | ||||
| Overtrunks | ||||
|
The archaea ( archaea , singular: archaeon ; from ancient Greek ἀρχαῖος archaĩos 'ancient', 'original'), formerly also called archaebacteria , archebacteria or primeval bacteria , form one of the three domains into which all cellular living beings are divided. The other two domains are the bacteria (Bacteria), which are combined with the archaea to form the prokaryotes , and the eukaryotes (eukaryotes), which, in contrast to the prokaryotes, have a cell nucleus .
description
Archaea are unicellular organisms that do not have a nucleus with a nuclear membrane like eukaryotic cells , but, like bacteria , self-contained DNA molecules ( circular chromosomes ) which are present in the cytoplasm as nuclear equivalent without a shell. Archaea and bacteria are therefore also called prokaryotes .
The separate position of the archaea as an independent domain is due to a number of genetic, physiological, structural and biochemical features, in particular significant differences in the sequence of the RNA contained in the ribosomes (the small ribosomal subunit, 16S rRNA ).
At the end of the 1970s, the American microbiologists Carl Woese and George Fox recognized and described the independence of archaea and their belonging to a separate systematic unit alongside bacteria ( eubacteria ) and eukaryotes. The researchers discovered striking differences from bacteria in the sequence of the ribosomal RNA. The structure of the cells and their peculiarities in metabolism also suggest a separate group of prokaryotes. These results were confirmed in the following years, and far-reaching advances in molecular biology made a general change in taxonomy necessary: eubacteria were renamed Bacteria and archaebacteria into Archaea . Both were described in 1990 as two separate domains in addition to the domain of the eukarya as part of a three-domain system . The Archaea are phylogenetically closer to the Eukarya than the Bacteria . Although no cell organelles can be found in archaea , they can develop special filaments comparable to a cytoskeleton to stabilize their shape .
Many cultivated archaea species are adapted to extreme environmental conditions. There are species that prefer to grow at temperatures above 80 ° C (hyper- thermophilic ), others live in highly concentrated salt solutions ( halophilic ) or in a strongly acidic environment (pH value below 0; acidophilic ) or a strongly basic environment ( pH value above 10; alkaliphilic ). Thermoplasmatales of the genus Picrophilus ( P. oshimae and P. torridus ) have a growth optimum at a pH of 0.7 and can even survive at a pH of −0.6.
Archaea are of interest in research because they may have retained features of early life on Earth. But their extraordinary metabolism is also of interest, for example the ability to grow at 110 ° C. The unusual metabolism is also interesting in terms of applications, for example archaea are used in soil and water remediation or for methane production in biogas plants .
So far, no pathogens from the archaea group are known.
properties
Archaea are more similar to eukaryotes than bacteria in terms of many molecular biological properties. Nevertheless, they have typical bacterial properties, e.g. B. the cell size, the lack of a cell nucleus , the type of cell division , they have a self-contained DNA molecule, also relatively simple locomotion organs ( flagella ) and, like bacteria, ribosomes with a sedimentation coefficient of 70S (although the archaeal ribosomes are more complex in their structure). The genes of both domains are organized in so-called operons . Archaea can also carry plasmids , such as an archaeon of the Crenarchaeota genus Sulfolobus .
The central molecular processes, for example translation and transcription , on the other hand, are quite similar to those of eukaryotes: Archaea use similar RNA polymerases ( rifampicin and streptolydigin resistant) composed of several protein subunits , and very similar initiation and elongation factors occur during translation The start of transcription is marked by a so-called TATA box .
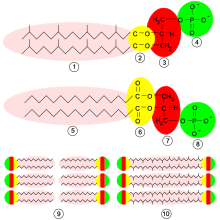
However, the archaea also have many unique properties, especially the structure of the cell wall shows clear differences to the other domains: The archaeal cell walls contain pseudopeptidoglycan (pseudomurein) and are generally very diverse in their structure: some archaea completely lack a cell wall ( thermoplasm ), others have highly complex cell walls consisting of many layers ( methanospirillum ). Due to their different structure, archaea are generally resistant to cell wall antibiotics. The composition of the archaeal plasma membrane differs: in bacteria and eukaryotes are fatty acids via an ester linkage to the glycerol molecules bound in archaea are found Glyceroldiether or even bis-glycerol tetraethers (single-layer membrane monolayer) and branched isoprene units instead of simple fatty acids. Hyperthermophilic archaea often have cell membranes (glycerol tetraether) that are stabilized in this way, which are not only more thermally stable, but can also adapt to acidic environments.
Some archaea species can move very quickly in relation to their size. With 400 to 500 "body lengths per second" (English " bodies per second ", abbreviated to "bps"), Methanocaldococcus jannaschii and Methanocaldococcus villosus are the fastest living beings measured so far. For comparison: A sports car with 400 bps would reach a speed of over 6000 km / h. The bacterium Escherichia coli , on the other hand, moves at around 20 bps, about as fast as a cheetah . In addition, archaea communicate with each other and thus coordinate their behavior.
Habitats
Most of the archaea known to date are extremophiles ; H. adapted to the extreme conditions of their biotopes . Many representatives have the ability to live at very high temperatures (hyperthermophiles over 80 ° C), very low or very high pH values ( acidophiles or alkali philes ), high salt concentrations ( halophiles ) or high pressures ( barophilia ).
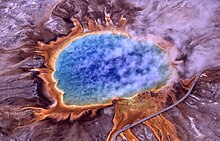
Hyperthermophilic archaea are often found in volcanic, marine ( black smoker ) and terrestrial ( geysers , solfatar fields ), e.g. B. volcanic habitats of Yellowstone National Park . Halophils thrive in high salinity environments, such as B. in the Dead Sea or in naturally occurring marine brines .
Methanogenic archaea are also “extreme” in a certain way: They only grow under anoxic conditions and often require molecular hydrogen for their metabolism . These archaea species are relatively widespread and occur in fresh water, sea and soil, but also as symbionts in the intestinal tract of animals and humans. Archaea have even been found in the folds of the human navel, although this is rare.
Because of this “extremophilia”, the ecological importance of the archaea was initially assessed as relatively minor. Only in recent years has it become clear through the use of finer molecular biological detection methods that archaea occur to a large extent in relatively cold seawater, but also in soils and freshwater biotopes. In certain oceanic areas z. B. Crenarchaeota up to 90% of the existing living things. Overall, it is estimated that around 1.3 × 10 28 archaea and 3.1 × 10 28 bacteria are found in the oceans .
The majority of the archaea species that are isolated and available as pure cultures in the laboratory are, however, still “extremophile”; in some cases cultivation has also been successful under less extreme conditions. Research to date shows that the archaea play an important role in the nitrogen , carbon and sulfur cycle in the earth's ecosystem.
metabolism
Most of the archaea species known to date are autotrophic , i. H. they do not need any organic substances for growth; they obtain the carbon to build up their body components exclusively through the assimilation of carbon dioxide . But heterotrophy , the extraction of carbon from organic compounds, is also widespread.
The majority of the archaea cultivated so far are characterized by an anaerobic metabolism; for many anaerobic archaea, oxygen (O 2 ) is toxic.
A special feature of archaeal metabolism is methanogenesis , which can only be accomplished by methane-producing archaea, the so-called methanogens . They have a number of unique cofactors , for example coenzyme F 420 or methanofuran .
Most hyperthermophilic archaea are anaerobes ; the energy-generating metabolism is either chemoorganotrophic or chemolithotrophic (the energy is obtained from chemical conversions of organic or inorganic compounds). Sulfur compounds often play a major role here: During the metabolism, the sulfur is reduced and energy is released in the process.
However, the sulfur metabolism of the extremely thermophilic and acidophilic species Acidianus ambivalens (previously: Desulfurolobus ambivalens ), from the order Sulfolobales , is known, which can aerobically oxidize sulfur.
Halophilic archaea are mostly aerobic-chemoorganotrophic, they get their energy from chemical reactions of organic compounds. Under anoxic conditions or when there is a lack of nutrients, many extreme halophiles are even capable of using light energy: They have the protein bacteriorhodopsin , which absorbs light and catalyzes the transfer of protons through the cytoplasmic membrane; the resulting electrochemical gradient drives the ATPase and thus the ATP synthesis.
morphology
Like the bacteria, the archaea are extremely diverse in shape. The sizes or lengths of the archaeal cells vary from about 0.4 ( Nanoarchaeum equitans ) up to 100 µm ( Methanospirillum hungatei ), on average the cells are about 1 µm in size. The cells show different shapes, e.g. E.g .: cocci (e.g. Methanococcus jannaschii ), rods ( Thermoproteus neutrophilus ), spirilla- shaped ( Methanospirillum hungatei ), lobed cocci ( Archaeoglobus fulgidus ), discs ( Thermodiscus maritimus ), long filaments ( Thermofilum pendens ) or even square ( Haloquadratum walsbyi ).
They often possess flagella ( flagella ) for locomotion, or thread-like appendages ( pili ) to attach to surfaces.
Systematics
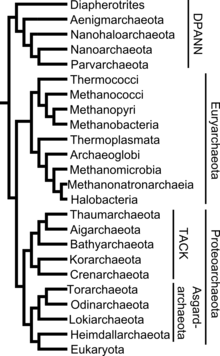
The placement of the archaea in the system of taxonomy is not entirely undisputed. Initially classified only by appearance and physiology, today, due to new possibilities, the classification by means of phylogenetic analysis, as proposed by Carl Woese (1977, 1990) , is generally accepted .
There is a fixed procedure for describing gender and species. The genus and species are determined by publication or validation in the International Journal of Systematic and Evolutionary Microbiology (IJSEM). Higher taxa can also be described here. The current status can be viewed in the List of Prokaryotic names with Standing in Nomenclature (LPSN), maintained by Jean Euzéby. This corresponds to the International Code of Nomenclature of Bacteria (ICNB). Taxa that do not conform to this standard are shown in quotation marks.
In addition, the global classification within the archaea and bacteria was reformed by means of phylogenetic analysis of the 16S rRNA gene. A current compilation of the taxa from this and numerous further publications appears in Bergey's Manual , and in Taxonomic Outline of the Bacteria and Archaea , whereby in addition to the 16S rRNA gene, further phylogenetic marker genes are now sometimes used. Some of these taxa are justified, but have not yet been published validly or are generally not recorded by the ICNB. Such taxa are shown in quotation marks.
The systematics presented here contains the taxa from phylum to family. There are contradicting entries for some taxa. These were checked for validity using original literature and a phylogenetic analysis.
A few years ago the description of the additional phyla " Korarchaeota " and " Nanoarchaeota " was published. A representative of the proposed phylum "Nanoarchaeota" was successfully co-cultivated and its genome sequenced, the so-called Nanoarchaeum equitans. There are enrichment cultures of the proposed phylum "Korarchaeota", initially detected using its 16S rRNA gene base sequences in samples from hot springs . From this the complete base sequence of the genome could now be published, given the informal name "Candidatus Korarchaeum cryptofilum ". Without isolated strains, the representatives of this phyla have no validated place in the taxonomy according to the current rules of the ICSB, but represent two of four known phyla of the archaea.
Superphylum " Euryarchaeota "
- Class Archaeoglobi
- Class Halobacteria
- Class Methanobacteria
- Class methanococci
- Class " Methanomicrobia "
- Class Methanopyri
- Class thermococci
- Class Thermoplasmata
Superphylum " DPANN "
- Phylum " Nanoarchaeota "
- Phylum " Nanohaloarchaeota "
- Super group " ARMAN "
- ...
Superphylum " Proteoarchaeota "
-
Super group " TACK " (Filarchaeota)
- Phylum " Aigarchaeota "
- Phylum " Bathyarchaeota "
- Phylum Crenarchaeota
- Class thermoprotei
- Phylum " Geoarchaeota "
- Phylum " Korarchaeota "
- Phylum Thaumarchaeota
- Class Nitrososphaeria
- Order Nitrososphaerales
- Family Nitrososphaeraceae
- Order Nitrososphaerales
- Class Nitrososphaeria
- Phylum Verstraetearchaeota
-
Supergroup " Asgard " (Asgardarchaeota)
- Phylum " Lokiarchaeota "
- Phylum " Helarchaeota "
- Phylum " Thorarchaeota "
- Phylum " Odinarchaeota "
- Phylum " Heimdallarchaeota "
Archaea in man
Archaea have been found in humans in the intestines ( colon ), in the mouth (dental flora), in the navel and in the vagina . Archaea, belonging to the genus Methanobrevibacter , especially Methanobrevibacter smithii , occur in the intestine . These belong to the methanogenic archaea . M. smithii is not found in the gut in all humans , and archaea have never been identified in infants under two years of age. In a study, archaea belonging to the phylum Thaumarchaeota were also found on the skin. Possibly the number of these archaea correlates with the frequency of sweating.
Methanogens of the species M. smithii and Methanosphaera stadtmanae live in association with syntrophic bacteria in the human digestive tract, so that they have an impact on digestion. These use the two products of bacterial fermentation hydrogen and formate for methanogenesis. A high concentration of hydrogen inhibits the ATP production of other bacteria. M. smithii also breaks down methane , which is toxic to humans. Therefore, the methanogens have a positive influence on the human intestinal flora . Whether these also influence how much energy humans can take in from food is still the subject of research.
Although archaea are in close contact with humans, there is no evidence of species pathogenic to humans . However, a correlation between the disease and the number of methanogenic archaea was demonstrated: the more archaea there were, for example in the (inflamed) gums , the more pronounced the corresponding periodontal disease . Archaea of the species Methanobrevibacter oralis occur in particular . The amount of methanogenic archaea was also increased in those areas in patients with colon cancer or diverticulosis . However, these archaea only contribute indirectly to the disease by promoting the growth of genuinely pathogenic bacteria - the archaea themselves are not. If archaea are viewed as “copathogens” or “ pathobionts ” (symbionts that become pathological under certain conditions), then the disease could be treated with medications that target these archaea. Statins, for example, inhibit the growth of Methanobrevibacter oralis, which is associated with periodontitis .
It is not yet clear why the known archaea are not pathogenic to humans. The uniqueness of many cofactors and vitamins that are not found in humans is not necessarily the cause. Even the fact that the microhabitats created by pathogenic processes are populated is not a unique selling point of the archaea - in principle, organisms with a similar (anaerobic, hydrogenotrophic) metabolism could also use these habitats.
Biotechnological potential
Archaeal metabolic functions, cell components or enzymes are used industrially. Especially the extremophiles have many properties that can be used biotechnologically. Some examples that are already in the development phase or application:
-
biotechnology
- Biogas production
- Microbial ore leaching (“microbial ore leaching” or “bioleaching”): In this process, low-grade, sulfidic ores are leached (the sulfide components are microbially oxidized to sulfate, thereby converting the heavy metals into a soluble state); this is used, for example, for the extraction of copper , zinc and nickel .
- Medicine: Use of cell wall components (so-called S-Layer ) as a carrier for vaccines
-
nanotechnology
- Use of the S-Layer for ultrafiltration
- Bacteriorhodopsin / purple membrane of phototrophic halophilic archaea as biosensors
- biology
- Obtaining heat-resistant enzymes , e.g. B. α- amylases , proteolytic enzymes, DNA polymerases
- Obtaining new restriction enzymes
literature
Archaea in standard works
- Georg Fuchs (ed.): General microbiology. (greeted by Hans G. Schlegel). 8th edition. Georg Thieme Verlag, Stuttgart / New York 2007, ISBN 978-3-13-444608-1 .
- Martin Dworkin, Stanley Falkow, Eugene Rosenberg, Karl-Heinz Schleifer, Erko Stackebrandt (Eds.) The Prokaryotes, A Handbook of the Biology of Bacteria . 7 volumes, 3rd edition. Springer-Verlag, New York a. a., 2006, ISBN 0-387-30740-0 .
- Joseph W. Lengeler, Gerhart Drews, Hans G. Schlegel (Eds.) Biology of the Prokaryotes . Georg Thieme Verlag, Stuttgart 1999, ISBN 3-13-108411-1 .
- H. König: Archaea. In: Encyclopedia of Life Sciences . John Wiley & Sons, 2003, ( doi: 10.1038 / npg.els.0000443 )
- Michael T. Madigan, John M. Martinko, Thomas Lazar (translator) and Freya Thomm-Reitz (translator): Brock Mikrobiologie . 11th, updated edition. Pearson Studies, 2009, ISBN 978-3-8273-7358-8 ;
Descriptions in the scientific literature
- CR Woese, O. Kandler , ML Wheelis: Towards a natural system of organisms: Proposal of the domains Archaea, Bacteria and Eucarya. In: Proc. Natl. Acad. Sci. UNITED STATES. 87 (12), 1990, pp. 4576-4579. PMID 2112744 ; PDF (free full text access)
- BM Karner, EF DeLong, DM Karl: Archaeal dominance in the mesopelagic zone of the Pacific ocean. In: Nature. Volume 409 (6819), 2001, pp. 507-510. PMID 11206545 ; doi: 10.1038 / 35054051
- R. Cavicchioli: Cold-adapted archaea. In: Nat Rev Microbiol . 4 (5), 2006, pp. 331-343. PMID 16715049 ; doi: 10.1038 / nrmicro1390
- H. Huber, MJ Hohn, R. Rachel, T. Fuchs, VC Wimmer, KO Stetter . A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. In: Nature. Volume 417 (6884), 2002, pp. 63-67. PMID 11986665 ; doi: 10.1038 / 417063a
- E. Conway de Macario, Alberto JL Macario: Methanogenic archaea in health and disease: a novel paradigm of microbial pathogenesis. In: Int J Med Microbiol. 299 (2), 2009, pp. 99-108. PMID 18757236 ; doi: 10.1016 / j.ijmm.2008.06.011
- R. Cavicchioli: Archaea - timeline of the third domain. In: Nat Rev Microbiol. 9 (1), 2011, pp. 51-61. PMID 21132019 ; doi: 10.1038 / nrmicro2482
- RE Valas, PE Bourne: The origin of a derived superkingdom: how a gram-positive bacterium crossed the desert to become an archaeon. In: Biology direct. Volume 6, 2011, p. 16, ISSN 1745-6150 . doi: 10.1186 / 1745-6150-6-16 . PMID 21356104 . PMC 3056875 (free full text).
- Hans-Peter Horz: Archaeal Lineages within the Human Microbiome: Absent, Rare or Elusive? In: Life (Basel). 5 (2), 2015, pp. 1333-1345. doi: 10.3390 / life5021333
Web links
Individual evidence
- ↑ a b Taxa above the rank of class. In: List of Prokaryotic names with Standing in Nomenclature . Retrieved August 8, 2017 .
- ^ Céline Brochier-Armanet, Bastien Boussau, Simonetta Gribaldo, Patrick Forterre: Mesophilic crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota . In: Nature Reviews Microbiology . tape 6 , no. 3 , 2008, p. 245-252 , doi : 10.1038 / nrmicro1852 , PMID 18274537 .
- ^ Merriam-Webster Online Dictionary
- ↑ AL Hartman et al. : The Complete Genome Sequence of Haloferax volcanii DS2, a Model Archaeon In: PLOS ONE . Vol. 5, No. 3, 2010, p. E9605 PMC 2841640 (free full text)
- ↑ JD Trent et al. : Chaperonin filaments: the archaeal cytoskeleton? In: PNAS. Vol. 94, no. 10, 1997, pp. 5383-5388. PMID 9144246 ; PDF (free full text access)
- ↑ F. Hara et al. : An actin homolog of the archaeon Thermoplasma acidophilum that retains the ancient characteristics of eukaryotic actin. In: J. Bacteriol. Vol. 189, no. 5, 2007, pp. 2039-2045. PMID 17189356 ; PDF (free full text access)
- ↑ C. Schleper et al. : A multicopy plasmid of the extremely thermophilic archaeon "Sulfolobus" effects its transfer to recipients by mating. In: J. Bacteriol. Vol. 177, 1995, pp. 4417-4426. PMID 7635827 .
- ↑ A. Pearson et al. : Factors Controlling the Distribution of Archaeal Tetraethers in Terrestrial Hot Springs. In: Appl. Environ. Microbiol. Vol. 74, 2008, pp. 3523–3532, as (PDF)
- ↑ E. Boyd et al. : The role of tetraether lipid composition in the adaptation of thermophilic archaea to acidity . In: Front Microbiol. tape 4 (62) , April 2013, doi : 10.3389 / fmicb.2013.00062 , PMC 3615187 (free full text).
-
^ Bastian Herzog, Reinhard Wirth: Swimming Behavior of Selected Species of Archaea. In: Applied and Environmental Microbiology. Volume 78, No. 6, 2012, pp. 1670–1674, doi: 10.1128 / AEM.06723-11
idw-online.de of February 28, 2012: Archaea are the fastest living beings in the world - microorganisms shine with a world record. - ↑ Guenther Witzany (Ed.): Biocommunication of Archaea . Springer, Switzerland 2017, ISBN 978-3-319-65535-2 , pp. 325 .
- ↑ Hulcr J, Latimer AM, Henley JB, Rountree NR, Fierer N, et al. (2012) A Jungle in There: Bacteria in Belly Buttons are Highly Diverse, but Predictable. PLoS ONE 7 (11): e47712. doi: 10.1371 / journal.pone.0047712
- ↑ T. Urich et al. : The sulfur oxygenase reductase from Acidianus ambivalens is a multimeric protein containing a low-potential mononuclear non-haem iron center. In: Biochem J. Volume 381, Part 1, 2004, pp. 137-146. doi: 10.1042 / BJ20040003
- ↑ CR Woese, GE Fox: Phylogenetic structure of the prokaryotic domain: the primary kingdoms. In: Proc. Natl. Acad. Sci. USA Vol. 74, No. 11, 1977, pp. 5088-5090. PMID 270744
- ↑ CR Woese et al. : Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. In: Proc. Natl. Acad. Sci. USA Volume 87, Issue 12, 1990, pp. 4576-4579. PMID 2112744
- ↑ BJ Tindall et al. : Valid publication of names of prokaryotes according to the rules of nomenclature: past history and current practice. In: Int J Syst Evol Microbiol. Volume 56, Issue 11, 2006, pp. 2715-2720. PMID 17082418 . doi: 10.1099 / ijs.0.64780-0 .
- ↑ International Journal of Systematic and Evolutionary Microbiology. (IJSEM)
- ↑ JP Euzéby: List of bacterial names with standing in nomenclature: a folder available on the Internet. In: Int. J. Syst. Bacteriol. Volume 47, 1997, pp. 590-592. PMID 9103655 - List of Prokaryotic names with Standing in Nomenclature ( Memento of December 30, 2010 in the Internet Archive ) (LPSN)
- ↑ S. P. Lapage et al. : International Code of Nomenclature of Bacteria, 1990 Revision . ( Bacteriological Code ). ASM Press, Washington, DC 1992, ISBN 1-55581-039-X .
- ↑ CR Woese et al. : A phylogenetic definition of the major eubacterial taxa. In: Syst. Appl. Microbiol. Volume 6, 1985, pp. 143-151. PMID 11542017 .
- ↑ Bergey's Manual Trust, Department of Microbiology, 527 Biological, Sciences Building, University of Georgia, Athens, GA 30602-2605, USA - Bergey's Manual
- ↑ George M. Garrity et al. : Taxonomic Outline of the Bacteria and Archaea . Release 7.7, March 6, 2007, Michigan State University Board of Trustees, www.taxonomicoutline.org
- ↑ C. Schleper, G. Jurgens, M. Jonuscheit M: Genomic studies of uncultivated archaea . In: Nat Rev Microbiol . 3, No. 6, 2005, pp. 479-88. doi : 10.1038 / nrmicro1159 . PMID 15931166 .
- ↑ W. Ludwig et al. : ARB: a software environment for sequence data. In: Nucleic Acids Res. Volume 32, 2004, pp. 1363-1371.
- ↑ E. Pruesse et al. : SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. In: Nuc. Acids Res. Volume 35, No. 21, 2007, pp. 7188-7196.
- ↑ P. Yarza et al. : The All-Species Living Tree Project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. In: Syst. Appl. Microbiol. 2008, doi: 10.1016 / j.syapm.2008.07.001 .
- ↑ H. Huber et al. : A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. In: Nature. 417, 2002, pp. 63-67. PMID 11986665 .
- ↑ E. Waters et al. : The genome of Nanoarchaeum equitans : insights into early archaeal evolution and derived parasitism. In: Proc Natl Acad Sci USA. 100, 2003, pp. 12984-12988. PMID 14566062 .
- ↑ SM Barns et al. : Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. In: Proc Natl Acad Sci US A. 93, 1996, pp. 9188-9193. PMID 8799176 .
- ↑ JG Elkins et al. : A korarchaeal genome reveals insights into the evolution of the Archaea. In: Proc Natl Acad Sci US A. 105, 2008, pp. 8102-8107. PMID 18535141 .
- ↑ P. De Vos, HG Trüper: Judicial Commission of the International Committee on Systematic Bacteriology. IXth International (IUMS) Congress of Bacteriology and Applied Microbiology. Minutes of the meetings, 14, 15 and 18 August 1999, Sydney, Australia. In: Int. J. Syst. Evol. Microbiol. Volume 50, 2000, pp. 2239-2244.
- ^ T. Cavalier-Smith: The neomuran revolution and phagotrophic origin of eukaryotes and cilia in the light of intracellular coevolution and a revised tree of life . In: Cold Spring Harb. Perspect. Biol . 6, No. 9, 2014, p. A016006. doi : 10.1101 / cshperspect.a016006 . PMID 25183828 . PMC 4142966 (free full text).
- ↑ Bojk A. Berghuis et al. : Hydrogenotrophic methanogenesis in archaeal phylum Verstraetearchaeota reveals the shared ancestry of all methanogens , in: PNAS 116 (11), March 12, 2019, pp. 5037-5044; doi: 10.1073 / pnas.1815631116
- ↑ Luis H. Orellana, T. Ben Francis, Karen Krüger, Hanno Teeling, Marie-Caroline Müller, Bernhard M. Fuchs, Konstantinos T. Konstantinidis, Rudolf I. Amann: Niche differentiation among annually recurrent coastal Marine Group II Euryarchaeota , in: Nature ISME Journal 13, pp. 3014-3036, August 26, 2019, doi: 10.1038 / s41396-019-0491-z , Fig. 1a
- ↑ Paul-Adrian Bulzu, Adrian-Stefan Andrei, Michaela M. Salcher, Maliheh Mehrshad, Keiichi Inoue, Hideki Kandori, Oded Beja, Rohit Ghai, Horia L. Banciu: Casting light on Asgardarchaeota metabolism in a sunlit microoxic niche , in: Nat Microbiol, Vol. 4, pp. 1129-1137, doi: 10.1038 / s41564-019-0404-y
- ↑ Kiley W. Seitz, Nina Dombrowski, Laura Eme, Anja Spang, Jonathan Lombard, Jessica R. Sieber, Andreas P. Teske, Thijs JG Ettema, Brett J Baker: Asgard archaea capable of anaerobic hydrocarbon cycling , in: Nature Communications , December 2019 doi : 10.1038 / s41467-019-09364-x
- ↑ Souri Somphanith: Plunging into the Unknown: Bellybutton Bacteria and You. In: PLOS Blogs .
- ^ E. Conway de Macario and Alberto JL Macario: Methanogenic archaea in health and disease: a novel paradigm of microbial pathogenesis. In: Int J Med Microbiol. 299 (2), 2009, pp. 99-108. PMID 18757236
- ^ A b Mor N. Lurie-Weinberger and Uri Gophna: Archaea in and on the Human Body: Health Implications and Future Directions . In: PLOS Pathogens . tape 11 , no. 6 , June 11, 2015, p. e1004833 , doi : 10.1371 / journal.ppat.1004833 , PMID 26066650 ( plos.org [accessed March 13, 2017]).
- ^ Joan L. Slonczewski, John W. Foster: Microbiology: A science with a future. 2nd Edition. Spektrum Akademischer Verlag, Berlin / Heidelberg 2012, ISBN 978-3-8274-2909-4 , p. 854.
- ↑ R. Cavicchioli et al. : Pathogenic archaea: do they exist? In: Bioessays. 25 (11), 2003, pp. 1119-1128. PMID 14579252 ; doi: 10.1002 / bies.10354
- ↑ a b Rustam I. Aminov: Role of archaea in human disease . In: Frontiers in Cellular and Infection Microbiology . tape 3 , January 1, 2013, doi : 10.3389 / fcimb.2013.00042 , PMID 23964350 ( frontiersin.org [accessed March 13, 2017]).
- ↑ W. Martin: Pathogenic archaebacteria: do they not exist Because archaebacteria use different vitamins? In: Bioessays. 26 (5), 2004, pp. 592-593. PMID 15112239 ; doi: 10.1002 / bies.20044 .
- ↑ R. Cavicchioli, P. Curmi: Response to William Martin's letter. In: Bioessays. 26 (5), 2004, p. 593. doi: 10.1002 / bies.20043 .