Mimivirus
| Mimivirus | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|

EM image of a virion (virus particle) of the |
||||||||||||||||||||||
| Systematics | ||||||||||||||||||||||
|
||||||||||||||||||||||
| Taxonomic characteristics | ||||||||||||||||||||||
|
||||||||||||||||||||||
| Scientific name | ||||||||||||||||||||||
| Acanthamoeba polyphaga mimivirus | ||||||||||||||||||||||
| Short name | ||||||||||||||||||||||
| APMV | ||||||||||||||||||||||
| Left | ||||||||||||||||||||||
|
Mimivirus is a genus of viruses belonging to the family mimiviridae , which amoebae act as natural hosts. With the family Mimiviridae , Mimivirus belongsto the giant viruses in the phylum of the Nucleocytoviricota (also English Nucleocytoplasmic large DNA viruses , NCLDV; original proposal “ Nucleocytoplasmaviricota ”; an even earlier proposal is that of an order “ Megavirales ” see left).
In the genus Mimivirus , as of March 2019, there is a single species Acanthamoeba polyphaga mimivirus ( APMV ) confirmed by the International Committee on Taxonomy of Viruses (ICTV ), which is therefore also the type species. In colloquial language and in older literature, APMV is usually referred to only as a mimivirus . However, there are a number of phylogenetically related large viruses suggested as additional members of this genus.
Discovery and history of research
The mimivirus APMV was discovered in 1992 during research work on Legionnaires' disease (Legionellosis) in an industrial cooling tower in Bradford (England), and it was found that it reproduces in the amoeba Acanthamoeba polyphaga . In 2003 it was identified at the Université de la Méditerranée in Marseille by a working group led by Didier Raoult . With a diameter of 400 nm , the virus particles ( virions ) of APMV are the size of small bacteria . Because of its size and its external resemblance to spherical bacteria ( cocci ), it was initially thought to be a gram-positive bacterium and was called Bradfordcoccus . When the mistake was recognized, the newly discovered virus was named mimicking virus , in allusion to its size and coloring properties . Eventually it became Mimivirus for short , with the name part mimi as an abbreviation for English mimicking microbe . In October 2004 Didier Raoult and colleagues published the structure of his genome in the journal Science . Incidentally, mimivirus was not the only virus to which this happened: Another example is the misannotated virus (suggested for Pithoviridae ), initially Amina Cherif Louazani, Sarah Aherfi, Rania Francis, Rodrigo Rodrigues, Ludmila Santos Silva, Dehia Sahmi, Said Mougari, Nisrine Chelkha , Meriem Bekliz, Lorena Silva, Felipe Assis, Fábio Dornas, Jacques Yaacoub Bou Khalil, Isabelle Pagnier, Christelle Desnues, Anthony Levasseur, Philippe Colson, Jônatas Abrahão, Bernard La Scola
The same team that discovered the APMV later discovered a slightly larger virus that Acanthamoeba castellanii mama virus (ACMV shortly Mama virus ) together with the Sputnik Virophagen that infects it. ACMV and APMV are so closely related that they are usually placed in the same genus, Mimivirus .
Until 2013, when an even larger virus, the Pandoravirus , was described, the viruses of the genus Mimivirus had the largest capsid diameter of all known viruses; in the meantime, however, it is also surpassed by close relatives Megavirus chilensis , Tupanvirus , Platanovirus , and other giant viruses such as the pithovirus .
Hosts
The first known mimivirus host is the amoeba Acanthamoeba polyphaga (genus Acanthamoeba , Amoebozoa ). So far only representatives of the genus Acanthamoeba , apart from A. polyphaga , A. castellanii and A. mauritaniensis , could be used as hosts of this virus in the laboratory , no cells from other unicellular or multicellular organisms. The natural hosts are unknown (as of 2015).
construction
The capsids of the virions (virus particles) of APMV appear hexagonal under an electron microscope , therefore the capsid geometry is icosahedral . There does not appear to be an outer viral envelope, suggesting that the virus does not exocytose the host cell .
The major protein of the Mimivirus -Kapsids consists of two domains of the sponge roller type ( English jellyroll fold ). This protein forms homotrimeric capsomeres as an organizational unit of the capsids. The capsomeres are packed hexagonally in the form of “daisies”: six capsomeres surround a recess between them.
Mimivirus virions have a capsid diameter of 400 nm .
Fibrils

B : AFM image of two detached surface fibers of the mimivirus .
C: Cryo-EM image of a mimivirus after partial degradation of the fibrils by means of bromelain .
D: AFM image of internal fibers of the mimivirus .
The capsid is covered with a compact layer of fibrils ( tegument ). The protein filaments (fibrils) protruding from the surface of the capsid have a length of about 100 nm (80–125 nm), bringing the total length of a virion to 600 nm. Deviations in the scientific literature make the numbers appear very inaccurate if for example the 'size' of the virion is sometimes reported as somewhere between 400 and 800 nm. Apart from differences between the individual virus strains in the genus Mimivirus , sometimes the total size is given with filaments, and sometimes the pure capsid diameter.
Investigations of these filaments by Klose et al. (2010) using an atomic force microscope found that these are often attached to a common support structure. However, at that time it could not be found out to which parts of the capsid surface these carriers are attached. Each fibril ends with a small spherical cap made of a protein with an unknown function. The protein filaments were found to be resistant to proteases unless they were treated with lysozyme . The filaments therefore appeared to be coated with peptidoglycan . That was all in good agreement with the fact that the Mimivirus can be stained by the Gram method .
With their strongly glycosylated surface, the filaments apparently play an important role in the approach to the host seedlings and the subsequent infection. The main component of the fibers is the protein R135 (alongside L725 and L829). Its structure resembles proteins from the family of glucose-methanol-choline oxidoreductases ( GMC oxidoreductases ), which have an N-terminal FAD binding domain and a C-terminal substrate recognition domain . The R135 closest homolog is an aryl alcohol - oxidase which the biological lignin degradation of plant cell walls is involved. Thus R135 could be involved in the perforation of the cell wall of its natural hosts, especially lignin-containing algae. Under laboratory conditions, however, none of the three proteins mentioned is absolutely necessary for infectivity .
Stargate

The pentagonal, star-shaped structure at one of the corners of the capsid, the so-called 'Stargate' (English for 'star gate', also called 'star structure' or 'starfish structure') is striking. If you look directly at this corner point (the center of the star), five triangular surfaces appear to lie between its rays. The beams are approximately 50 nm wide, 40 nm thick, and 200 nm long; they almost reach the neighboring corner points of the icosahedral capsids. The Stargate is not covered by fibrils. The presence of this structure changes the geometry of the capsid by deviating its geometry from the ideal icosahedral shape: in fact, on closer inspection, only a single axis with five-ray symmetry runs through the virion, which runs through the center of the star (called the vertex).
The symmetry of the capsid is indicated differently with a triangulation number T = 972–1141 or T = 1200.
Since no hexagonally arranged depressions can be observed on the surface of the star structure, it is assumed that this is a protein that differs from the main capsid protein.
The Stargate plays a special role in the infection of the host cell: During the infection, the "lock" at the apex opens and the viral core (with DNA and prefabricated proteins) is released from the capsid into the cytosol of the host cell (via phagocytosis ) . That is the reason why the star structure as "Stargate" ( English stargate is called).
Nucleocapsid
The mimivirus shares several morphological features with all members of the NCLDV virus group. Immediately under the capsid of the mimivirus , for example, there are two electron-dense layers that are interpreted as membranes. Underneath these membranes is an approximately 7 nm thick protein shell, in which the linear double-stranded DNA of the virus is enclosed. This condensed central nucleus of the virion appears under the electron microscope as a dark area, the so-called 'nucleocapsid'. This area contains the large genome of the virus, as well as mRNAs and pre-made proteins . Since all other NCLDVs have an internal lipid layer that surrounds the central core, this is also believed to be the case with mimivirus . The walls of the nucleocapsid are about 30 nm behind the walls of the capsid, in the area of the star structure (the stargate) the surface of the nucleocapsid is additionally lowered. It is believed that the space between the tip of the star structure and the nucleocapsid is filled with hydrolytic enzymes that are necessary for the virus to penetrate the host cell. Internal protein strands were discovered between the capsid and the nucleocapsid, which apparently stabilize the mutual spatial positioning of the two parts.
Genome
Compared to most other viruses, APMV has a large and complex genome that consists of a single linear DNA double strand (dsDNA). The genome length of the APMV wild type (starting variant Mimivirus M1) was determined by Raoult et al. (2004) given as 1,181,404 bp , this value was given by Bäckström et al. (2019) corrected slightly to 1,181,594 bp. This corresponds to about 800 nm. The fiberless variant Mimivirus M4 has only 0.993 Mbp, between M2 with 1.10 Mbp and M3 with 1.10 Mbp. The GC content of APMV is 28%. There are at APMV 1260 Open reading frames (ORFs English open reading frames ), including predicted 979 coding genes. This goes well beyond the minimum set of 4 genes that are required for a virus, such as phages MS2 and Qβ . Detailed studies of the genome repeatedly correct sequence errors and possibly discover new reading frames.
The proportion of non-coding DNA is therefore only about 9.5 to 10%. Open reading frames are separated by gaps of approximately 157 nucleotide pairs. Two DNA segments with the designation R ( English right ) and L ( English left ) encode approximately the same number of genes (450 and 465, respectively, according to data from 2010). The GC content is low at 28%. In the vicinity of the ends of the DNA molecule inverted repeats' (were English inverted repeats ) found with 617 nucleotide pairs. It is believed that the mutual interaction of these sites can lead to the formation of a Q-structure - circular DNA with two small appendages.
During the analysis, at least 21 genes with homology to known proteins were found, including those that were previously not known from any other virus, but only from cellular organisms, including aminoacyl-tRNA synthetase . 43 genes are homologous to those of other giant viruses (NCLDVs). Like other giant viruses, Mimivirus contains several genes for sugar, lipid and amino acid metabolism. There were also metabolic genes that had not previously been found in any other virus.
From purified virions were several mRNA - transcripts are obtained. As with other NCLDVs, transcripts for DNA polymerase , a capsid protein and a TFII-like transcription factor were found. However, three different aminoacyl-tRNA synthetase transcripts and four unknown mRNA molecules that are specific for the mimivirus were also found. These prepackaged transcripts can be translated without viral gene expression and are likely required for mimivirus replication . Other DNA viruses, such as human cytomegalovirus and herpes simplex virus type 1, also contain packaged mRNA transcripts.
Mimivirus is one of the few dsDNA viruses in whose genome an intein-coding sequence has been detected. Inteins are protein domains that catalyze their own removal from a carrier molecule and the subsequent linkage of the ends formed . Such a sequence is present in the mimivirus gene for DNA polymerase B.
Due to the extraordinarily complex genetic makeup of the virus, the question arises for some researchers where the boundary between animate and inanimate nature runs, i.e. how " living beings " are to be defined.
Propagation cycle
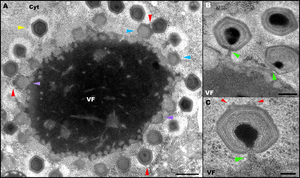
A) TEM image of an intracellular virus factory, with virus particles (virions) of Mimivirus in different stages of assembly
- VF: virus factory (viroplasma)
- Cyt: cytoplasm
- violet arrows: empty, still fiberless virions in the initial stage of the Composition, occurring in close proximity to the periphery of the virus factory
- blue arrows: partially mounted empty, fiber-free virions
- yellow arrows: mature, fiber-covered virions that are now further away from the virus factory than the immature particles
- red arrows: when there are several virus particles A 'Stargate', which is regularly located at the distal point of the factory, can be seen
B) and C) TEM image of the mimivirus particles
- green arrows: Mimivirus particles in the 'DNA packaging' stage (DNA packaging)
- red arrows: two edges of a stargate opposite the location of the DNA packaging
The details and various stages in the mimivirus replication cycle , such as apparent cell surface binding and entry, viral nucleus release, DNA replication, transcription, translation, and finally daughter assembly and release -Virions, are not yet well known. However, the scientists created the general overview given above using electron microscope images of infected cells. All stages of the reproduction cycle take place in the host cell's cytoplasm .
The infection of the amoeba with a mimivirus probably occurs according to the following scenario:
- The Mimivirus virions are similar in size and the presence of characteristic polysaccharides on the surface of bacteria (see Gram stain , name). They are therefore absorbed by the amoeba as food during an endocytosis process . The polysaccharides act as a chemical receptor and initiate the accumulation. As a result of endocytosis, the virions reside in endosomes within the cell.
- The protein filaments are partially lysed in the endosomes , which allows the capsid to interact with the endosome membrane.
- ~ 2 hours after infection: The capsid opens in the area of the star structure (Stargate), the inner membrane fuses with the endosome membrane and the contents of the capsid are released into the cytoplasm.
- After the core particle (the inner part of the nucleocapsid) has passed into the cytoplasm, the synthesis of the viral mRNA begins due to the presence of the viral transcription apparatus . These mRNAs accumulate inside the core particle in the form of granules. From the outside, the virus seems to have disappeared and everything in the cell looks normal (dark phase, English eclipse phase ).
- 4-5 hours after infection: The viral DNA leaves the core particle and is unpacked so that replication can begin. As a result, a so-called “ virus factory ” is created next to the empty shell of the core particle - a place for the synthesis of the individual components of the virions and their subsequent assembly. When several virus particles have entered the cell, the “factories” they have formed merge into one as they grow. You can now see small accumulations in some areas of the cell.
- 6–9 hours after infection: Composition (assembly) of the capsids with simultaneous DNA packaging on the periphery of the “virus factories”. An unusual property of the mimivirus is that DNA is packed. The mimivirus virions are clearly visible in the cell.
- 14-24 hours after infection: the amoebic cells are lysed, i.e. H. they burst and the virions are released. In this way, more than 300 units are produced per host cell.
The transmission occurs through passive diffusion .
Possible pathogenicity
It has been speculated that mimivirus could cause certain types of pneumonia . This is mainly based on indirect evidence in the form of antibodies against the virus found in pneumonia patients. Due to the few publications to date, however, it is currently difficult to classify the mimivirus as a possible pathogen. A large proportion of pneumonia cases have no apparent cause. A mimivirus has been isolated from a Tunisian woman suffering from pneumonia, and there is evidence from cell cultures that mimivirus can infect macrophages and is replicated in them. Thus, under experimental conditions, it has been observed that mimivirus can infect human macrophages; H. can penetrate the cells via phagocytosis and replicate there. In addition, antibodies to the mimivirus have been found in a small number of patients with pneumonia in several studies. An isolated case of pneumonia by a laboratory assistant working with cultures of this virus has also been described. The level of antibodies against mimivirus in his blood was also increased. However, the presence of antibodies to the virus in itself is not an indication of its pathogenicity. It is possible that the mimivirus simply has strong immunogenic properties; H. triggers a clear immune response. Also, in none of the registered cases was it possible to isolate the virus in its pure form from samples of fluids obtained from patients.
The fiber proteins R135 and L829 have been identified as the main antigens of the mimivirus : however, the fiberless variant mimivirus M4 showed no reactivity with sera from human patients, which confirms that these proteins are absent in M4.
Resistance property
The Zamilon virus is a satellite virus that Mimiviren lines B and C affects, but not the Mimiviren line A. This fact have a MIMIVIRE, English Mimivirus virophage resistance element , called resistance , which works similarly to the CRISPR / Caspari System .
It has also been shown that there is not only a gene transfer between the amoeboid hosts and giant viruses as intracellular viral endocytobionts (organisms that live or multiply in the cells of other organisms) but even between the viruses and bacterial endocytobionts that are present at the same time.
Features of the Mimiviridae in comparison
| virus | Aminoacyl-tRNA synthetase | Octocorallia -like MutS | Protein filaments (length) | Stargate | Well-known virophage | Cytoplasmic Virion Factory |
host |
|---|---|---|---|---|---|---|---|
| Megavirus chilensis | 7 ( Tyr , Arg , Met , Cys , Trp , Asn , Ile ) | Yes | yes (75 nm) | Yes | No | Yes | Acanthamoeba ( Unikonta , Amoebozoa ) |
| Mamavirus ACMV | 4 (Tyr, Arg, Met, Cys) | Yes | yes (120 nm) | Yes | Yes | Yes | Acanthamoeba (Unikonta, Amoebozoa) |
|
Mimivirus ApMV (wild type M1) |
4 (Tyr, Arg, Met, Cys) | Yes | yes (120 nm) | Yes | Yes | Yes | Acanthamoeba (Unikonta, Amoebozoa) |
| Mimivirus M4 (soon / fiberless variant) |
2 (Met, Cys) | No | No | Yes | resistant | Yes | Acanthamoeba (Unikonta, Amoebozoa) |
| Cafeteria roenbergensis virus | 1 (Ile) | Yes | No | No | Yes | Yes | Phagotrophic protozoa ( heteroconta , stramenopiles ) |
Systematics
External system
The genus Mimivirus and some others - not yet confirmed by the ICTV as of March 2019 - genetically similar genera and species of the family Mimiviridae (such as " Mamavirus ", " Megavirus " and " Moumouvirus ") form a clade of mimiviruses designated as Group I in a broader sense. For these it was suggested to raise them as " Megamimivirinae ", " Megavirinae " or " Mimivirinae " to the rank of a subfamily and thus to differentiate them from other subfamilies of the Mimiviridae, which are also proposed , namely:
- Cafeteria virus group (group II)
- Klosneuvirus group - as a possible subfamily " Klosneuvirinae "
Within the Mimiviridae group I the following lines stand out:
- Line A: Mimivirus group : Mimiviruses in the narrower sense
- Line B: Moumouvirus group (Moumouviren)
- Line C: Courdo11 group with " Megavirus chilensis " (species, including Courdo11 virus)
- Tupan virus group (Tupan viruses)
Since so far (March 2019) only the genus Mimivirus has been confirmed by the ICTV , it remains open at the moment which candidates for this proposed subfamily will be assigned to this genus and for which separate genera are to be set up. It seems certain, however, that the representatives of line A belong in each case to the genus Mimivirus .
The Wilson et al. Candidate described in 2017 " gvSAG AB-566-O17 " (designated by the NCBI as the species " Mimivirus AB-566-O17 ") is, according to the authors (Fig. 2), more widely related to APMV than CroV (but more closely as a representative of the OLPG group ). It can therefore not be assigned to any of the above groups (in particular not to the genus Mimivirus ).
Internal system
Systematics of line A (mimiviruses in the narrower sense):
- Genus Mimivirus
-
- Species: Acanthamoeba polyphaga mimivirus (APMV) (confirmed by ICTV as of March 2018) - Location: Bradford , England, UK (informally Mimivirus see s. )
- Mimivirus M1 ( English wild type ) - wild type (natural variant of origin) from Bradford
- Mimivirus M1 and M2 - intermediate forms between M1 and M4 with shorter fibrils than the wild type
- Mimivirus M4 ( english soon / fiberless variant ) - fiber-free variant is not from Sputnik Virophagen infested
- Mimivirus Bombay (MVB, alias Bombay virus) - Discovery site: Mumbai , India
- Mimivirus shirakomae (alias Shirakomae virus) - Discovery site: Shirakoma Pond , Nagano , Japan
- Mimivirus kasaii (aka Kasaii virus) - Found in: Arakawa (river), Tokyo , Japan
- Samba virus - location: Rio Negro , Brazil
- Mimivirus amazonia (aka Amazonia virus, Amazonian virus) - Location: Rio Negro, Brazil
- Oyster virus - Location: Florianópolis , Brazil
- Kroon Virus - Location: Lagoa Santa, Brazil
- The NCBI still cites the Saudi moumouvirus (SDMV) under APMV, but itself refers to the work of Bajrai et al. (2016), which correctly refers to group d in the title " Saudi Moumouvirus, the First Group B Mimivirus Isolated from Asia ". H. Line B ( Moumouvirus group) refers.
- Species: " Acanthamoeba castellanii mamavirus " (ACMV, alias Acanthamoeba castellanii mimivirus ) - Location: Paris, France
- Species: “ Hirudovirus ” - Discovery site: Tunisia
- Hirudovirus sangsue
- Species: " Niemeyer virus " - Location: Belo Horizonte , Brazil
- Species: " Mimivirus battle6 "
- Species: " Mimivirus battle7 "
- Species: " Mimivirus battle19 "
- Species: " Mimivirus battle27 "
- Species: " Mimivirus battle57 "
- Species: " Mimivirus battle66 "
- Species: " Mimivirus battle83 "
- Species: " Mimivirus battle86 "
- Species: " Mimivirus Cher "
- Species: " Mimivirus dakar4 "
- Species: Mimivirus fauteuil (alias " Fauteuil virus ") - cf. Fauteuil virus FD
- Species: " Mimivirus huitre A06 "
- Species: " Mimivirus lactour " (alias " Mimivirus lactours ", Lactours virus ) - cf. Lactours virus LT2
- Species: " Mimivirus lentille " (alias " Lentille virus ", " Acanthamoeba polyphaga lentillevirus ") - cf. Lentille virus CL
- Species: " Mimivirus longchamps " (alias " Longchamps virus ") - cf. Longchamps virus FPL
- Species: " Mimivirus marais " (aka " Marais virus ")
- Species: " Mimivirus montadette2 "
- Species: " Mimivirus pointerouge1 " - cf. Pointerouge virus 1 (aka Pointe-Rouge 1 virus ) - Location: Marseille , France
- Species: " Mimivirus pointerouge2 " - cf. Pointerouge virus 2 (alias Pointe-Rouge 2 virus ) - Discovery site: Marseille, France
- Species: Mimivirus SR1 (alias " Mimivirus-like virus SR1 ") - Location: Serendah village, Malay Peninsula - waterfall
- Species: Mimivirus SR4 (alias " Mimivirus-like virus SR4 ") - Discovery site: Serendah village, Malay Peninsula - middle between waterfall and village
- Species: " Mimivirus SR9 " (alias " Mimivirus-like virus SR9 ") - Discovery site: Serendah village, Malay Peninsula - mouth of the tributary into the lake
- Species: " Mimivirus T2 "
- Species: " Mimivirus T3 "
- Species: " Mimivirus terra2 " (alias Terra2 virus ) - Discovery site: Marseille, France
- Terravirus2 TAO-TJA
- Species: " Mimivirus univirus "
Cladogram
Proposed phylogenetic tree of the genus Mimivirus according to Abrahão et al. (2018), Fig. 4:
| Mimivirus |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
Note: Italics for suggested species.
literature
- Stefanie Reinberger: Revolution of the giant viruses . In: Spectrum of Science . Spektrum der Wissenschaft Verlagsgesellschaft , Heidelberg, May 2012, pp. 14–16.
Web links
- Jean-Michel Claverie (Ed.): Images of the Mimivirus , by: Structural & Genomic Information Laboratory CNRS UPR, Marseille
- NCBI: Reference sequence NC_006450
- David R. Wessner: Discovery of the Giant Mimivirus , on: Nature Masterclasses
- Philippe Colson, Bernard La Scola, Anthony Levasseur, Gustavo Caetano-Anollés, & Didier Raoult: Mimivirus: leading the way in the discovery of giant viruses of amoebae , in: Nature Reviews Microbiology, Volume 15, pp. 243-254, 27. February 2017, doi: 10.1038 / nrmicro.2016.197
- Mimivirus - an overview , on: ScienceDirect, 2012
- Scinexx: Giant Viruses on the Edge of Life , February 28, 2018
- Andreas Jahn: Das Virus-Virus , on: Spektrum.de from August 6, 2008
- Biology side: Mimivirus
- Iddo: Size matters. Life is live. , on: Byte Size Biology, blog from May 1st, 2009, with picture of the opening Stargate, identical to Zaubermann et al. (2008), Fig. 5.
- Adrian De Novato: New study shines light on mysterious giant viruses , on: phys.org from May 8, 2020 (the picture apparently shows a typical representative of the genus Mimivirus with Stargate (opening for DNA) and Tegument (shell made of fibrils).
- Graziel Oliveira, Bernard La Scola, Jônatas Abrahão: Giant virus vs amoeba: fight for supremacy , in: Virol J 16, 126, November 4, 2019, doi: 10.1186 / s12985-019-1244-3 , PDF
Individual evidence
- ↑ a b c d e f g h i j k l m n o p q r s t u v Jan Diesend, Janis Kruse, Monica Hagedorn, Christian Hammann: Amoebae, Giant Viruses, and Virophages Make Up a Complex, Multilayered Threesome , in: Frontiers in Cellular and Infection Microbiology 7, January 2018, doi: 10.3389 / fcimb.2017.00527 , via ResearchGate , Fig. 1 (NCLDVs and ' Megavirales ' are not correctly referred to as' families' in this work, what is meant is' groups ').
- ↑ a b c d e ICTV: ICTV Taxonomy history: Acanthamoeba polyphaga mimivirus , EC 51, Berlin, Germany, July 2019; Email ratification March 2020 (MSL # 35)
- ↑ a b Christoph M. Deeg, Cheryl-Emiliane T. Chow, Curtis A. Suttle: The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea ... , in: eLife Sciences 7, March 2018, doi: 10.7554 / eLife.33014
- ↑ a b c Center national de la recherche scientifique: List of the main “giant” viruses known as of today (March 2019) , Université Aix Marseille, March 2019.
- ↑ a b Frederik Schulz, Lauren Alteio, Danielle Goudeau, Elizabeth M. Ryan, Feiqiao B. Yu, Rex R. Malmstrom, Jeffrey Blanchard, Tanja Woyke: Hidden diversity of soil giant viruses , in: Nature Communicationsvolume 9, Article number: 4881 (2018) of November 19, 2018, doi: 10.1038 / s41467-018-07335-2
- ↑ a b c ViralZone: Mimivirus . ExPASy. Retrieved July 8, 2019.
- ↑ ICTV : Master Species List 2018b.v2 . Retrieved August 6, 2019. MSL # 34v
- ^ A b Bernard La Scola, S. Audic, C. Robert, L. Jungang, X. de Lamballerie, M. Drancourt, R. Birtles, JM Claverie, D. Raoult: A giant virus in amoebae. In: Science. 299, 2003, p. 2033. PMID 12663918
- ↑ Laurie O'Keefe: Sizing Up Viruses , on: The Scientist. Illustration for Didier Raoult: Viruses Reconsidered , ibid 28 February 2014
- ↑ a b c d Didier Raoult, S. Audic, C. Robert, C. Abergel, P. Renesto, H. Ogata, B. La Scola, M. Suzan, JM Claverie: The 1.2-megabase genome sequence of Mimivirus , in : Science 306 (5700), November 19, 2004, pp. 1344-1350, doi: 10.1126 / science.1101485 , PMID 15486256
- ↑ Discovery and Further Studies on Giant Viruses at the IHU Mediterranee Infection That Modified the Perception of the Virosphere , in: Viruses 11 (4), March / April 2019, pii: E312, doi: 10.3390 / v11040312 , PMC 6520786 (free full text) , PMID 30935049
- ^ H. Pearson: 'Virophage' suggests viruses are alive . In: Nature . 454, No. 7205, 2008, ISSN 0028-0836 , pp. 677-677. bibcode : 2008Natur.454..677P . doi : 10.1038 / 454677a .
- ↑ a b c d e Suzan-Monti M, La Scola B, Raoult D: Genomic and evolutionary aspects of Mimivirus . In: Virus Research . 117, No. 1, April 2006, pp. 145-155. doi : 10.1016 / j.virusres.2005.07.011 . PMID 16181700 .
- ↑ a b c d e f g h i j k Thomas Klose, Dominik A. Herbst, Hanyu Zhu, Joann P. Max, Hilkka I. Kenttämaa, Michael G. Rossmann: A Mimivirus Enzyme that Participates in Viral Entry , in: Structure Volume 23, No. 6, June 2, 2015, pp. 1058-1065, doi: 10.1016 / j.str.2015.03.023
- ↑ a b c d e f g h Xiao C., Kuznetsov YG, Sun S., Hafenstein SL, Kostyuchenko VA, Chipman PR, Suzan-Monti M., Raoult D., McPherson A., Rossmann MG: Structural studies of the giant mimivirus . In: PLoS Biol . 7, No. 4, 2009, p. E92. doi : 10.1371 / journal.pbio.1000092 . PMID 19402750 . PMC 2671561 (free full text).
- ↑ Jean-Michel Claverie, Chantal Abergel, Hiroyuki Ogata: Mimivirus . In: Curr Top Microbiol Immunol . 328, 2009, pp. 89-121. doi : 10.1007 / 978-3-540-68618-7_3 . PMID 19216436 .
- ↑ a b c d Klose Thomas, Kuznetsov YG, Xiao C., Sun S., McPherson A., Rossmann MG: The three-dimensional structure of Mimivirus . In: Intervirology . 53, No. 5, 2010, pp. 268-273. doi : 10.1159 / 000312911 . PMID 20551678 . PMC 2895761 (free full text).
- ↑ see also vanillyl alcohol oxidase
- ↑ In physics, this geometrical principle is known as symmetry breaking (breaking a discrete symmetry) .
- ↑ a b c d e Author = Nathan Zauberman, Y. Mutsafi, DB Halevy, E. Shimoni, E. Klein, C. Xiao, S. Sun, A. Minsky: Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus . In: PLoS Biol . 6, No. 5, 2008, p. E114. doi : 10.1371 / journal.pbio.0060114 . PMID 18479185 . PMC 2430901 (free full text).
- ↑ a b Disa Bäckström, Natalya Yutin, Steffen L. Jørgensen, Jennah Dharamshi, Felix Homa, Katarzyna Zaremba-Niedwiedzka, Anja Spang, Yuri I. Wolf, Eugene V. Koonin, Thijs JG Ettema; Richard P. Novick (Ed.): Virus Genomes from Deep Sea Sediments Expand the Ocean Megavirome and Support Independent Origins of Viral Gigantism , in: mBio Vol. 10, No. 2, March – April 2019, pp. E02497-18, PDF , doi: 10.1128 / mBio.02497-18 , PMC 6401483 (free full text), PMID 30837339 , ResearchGate
- ↑ a b c d e Mickaël Boyer, Saïd Azza, Lina Barrassi, Thomas Klose, A. Campocasso, I. Pagnier, G. Fournous, A. Borg, C. Robert, X. Zhang, C. Desnues, B. Henrissat, MG Rossmann, B. La Scola, D. Raoult: Mimivirus shows dramatic genome reduction after intraamoebal culture , in: Proc Natl Acad Sci (PNAS) USA 108 (25), June 21, 2011, pp. 10296-10301, doi: 10.1073 /pnas.1101118108 , PMID 21646533 , PMC 3121840 (free full text)
- ↑ David M. Needham, Alexandra Z. Worden et al .: A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators , in: PNAS, 23 September 2019, doi: 10.1073 / pnas.1907517116 , ISSN 0027-8424 , here: Supplement 1 (xlsx)
- ↑ Lansing M. Prescott: 2nd edition (Ed.): Microbiology . Wm. C. Brown Publishers, Dubuque, IA 1993, ISBN 0-697-01372-3 .
- ↑ a b c d Jean-Michel Claverie, Chantal Abergel: Mimivirus and its virophage . In: Annu Rev Genet . 43, No. 49-66, 2009. doi : 10.1146 / annurev-genet-102108-134255 . PMID 19653859 .
- ↑ Jean-Michel Claverie, Hiroyuki Ogata, Stéphane Audic, Chantal Abergel, Pierre-Edouard Fournier, Karsten Suhre: Mimivirus and the emerging concept of 'giant' virus . In: Virus Research . 117, No. 1, 2006, pp. 133-144. arxiv : q-bio / 0506007 . doi : 10.1016 / j.virusres.2006.01.008 . PMID 16469402 .
- ↑ Hiroyuki Ogata, Didier Raoult, Jean-Michel Claverie: A new example of viral intein in Mimivirus . In: Virol J . 2, No. 8, 2005. PMID 15707490 . PMC 549080 (free full text).
- ↑ a b Mutsafi Y., Zauberman N., Sabanay I., Minsky A .: Vaccinia-like cytoplasmic replication of the giant Mimivirus . In: Proc Natl Acad Sci USA (PNAS) . 107, No. 13, 2010, pp. 5978-5982. doi : 10.1073 / pnas.0912737107 . PMID 20231474 . PMC 2851855 (free full text).
- ↑ a b La Scola B., Marrie TJ, Auffray JP, Raoult D .: Mimivirus in pneumonia patients . In: Emerg Infect Dis. . 11, No. 3, 2005, pp. 449-452. PMID 1575756 3. PMC 3298252 (free full text).
- ↑ Marrie TJ, Durant H, Yates L: Community-Acquired Pneumonia Requiring Hospitalization: 5-Year Prospective Study . In: Reviews of Infectious Diseases . 11, No. 4, 1989, pp. 586-99. doi : 10.1093 / clinids / 11.4.586 . PMID 2772465 .
- ↑ Saadi H, Pagnier I, Colson P, Cherif JK, Beji M, Boughalmi M, Azza S, Armstrong N, Robert C, Fournous G, La Scola B, Raoult D: First isolation of Mimivirus in a patient with pneumonia . In: Clinical Infectious Diseases . 57, No. 4, August 2013, pp. E127–34. doi : 10.1093 / cid / cit354 . PMID 23709652 .
- ↑ a b E. Ghigo, J. Kartenbeck, P. Lien, L. Pelkmans, C. Capo, JL Mege, D. Raoult: Ameobal pathogen mimivirus infects macrophages through phagocytosis. In: PLoS pathogens. Volume 4, number 6, June 2008, p. E1000087, doi: 10.1371 / journal.ppat.1000087 , PMID 18551172 , PMC 2398789 (free full text).
- ↑ Vincent A., La Scola B., Papazian L .: Advances in Mimivirus pathogenicity . In: Intervirology . 53, No. 5, 2010, pp. 304-309. doi : 10.1159 / 000312915 . PMID 20551682 .
- ↑ Berger P., Papazian L., Drancourt M., La Scola B., Auffray JP, Raoult D .: Ameba-associated microorganisms and diagnosis of nosocomial pneumonia . In: Emerg Infect Dis . 12, No. 2, 2006, pp. 248-255. PMID 16494750 . PMC 3373093 (free full text).
- ^ Raoult D., Renesto P., Brouqui P .: Laboratory infection of a technician by mimivirus . In: Ann Intern Med . 144, No. 9, 2006, pp. 702-703. PMID 16670147 .
- ↑ Vanspauwen MJ et al. : Infections with mimivirus in patients with chronic obstructive pulmonary disease . In: Respiratory Medicine . 106, No. 12, 2012, pp. 1690-1694. doi : 10.1016 / j.rmed.2012.08.019 .
- ↑ Anthony Levasseur, Meriem Bekliz, Eric Chabrière, Pierre Pontarotti, Bernard La Scola, Didier Raoult: MIMIVIRE is a defense system in Mimivirus did confers resistance to virophage. In: Nature , 2016, doi: 10.1038 / nature17146 .
- ↑ Ewen Callaway: CRISPR-like 'immune' system discovered in giant virus , in: Nature : News, June 29, 2016
- ↑ Nisrine Chelkha, Anthony Levasseur, Pierre Pontarotti, Didier Raoult et al. : A Phylogenomic Study of Acanthamoeba polyphaga Draft Genome Sequences Suggests Genetic Exchanges With Giant Viruses , in: Frontiers in Microbiology 9: 2098, September 2018, doi: 10.3389 / fmicb.2018.02098
- ↑ Patrick L. Scheid: Free-Living Amoebae and Their Multiple Impacts on Environmental Health , in: Reference Module in Earth Systems and Environmental Sciences, February 27, 2018, doi: 10.1016 / B978-0-12-409548-9.10969-8 , here: text following Fig. 8 (right column)
- ↑ A homology between CRISPR and MIMIVIRE does not seem impossible, but certainly needs further investigations.
- ^ MG Fischer, CA Suttle: A Virophage at the Origin of Large DNA Transposons . In: Science . tape 332 , no. 6026 , 2011, pp. 231-234 , doi : 10.1126 / science.1199412 , PMID 21385722 .
- ↑ NCBI: Mimivirus AB-566-O17 (acronym: gvSAG AB-566-O17) (species)
- ↑ William H Wilson, Ilana C Gilg, Mohammad Moniruzzaman, Erin K Field, Sergey Koren, Gary R LeCleir, Joaquín Martínez Martínez, Nicole J Poulton, Brandon K Swan, Ramunas Stepanauskas, Steven W Wilhelm: Genomic exploration of individual giant ocean viruses , in: ISME Journal 11 (8), August 2017, pp. 1736–1745, doi: 10.1038 / ismej.2017.61 , PMC 5520044 (free full text), PMID 28498373
- ↑ a b c NCBI: Acanthamoeba polyphaga mimivirus (species)
- ↑ Clara Rolland, Julien Andreani, Amina Cherif Louazani, Sarah Aherfi, Rania Francis, Rodrigo Rodrigues, Ludmila Santos Silva, Dehia Sahmi, Said Mougari, Nisrine Chelkha, Meriem Bekliz, Lorena Silva, Felipe Assis, Fábio Dornas, Jacques Yaacoub Bou Khalil, Isabelle Pagnier, Christelle Desnues, Anthony Levasseur, Philippe Colson, Jônatas Abrahão, Bernard La Scola: Discovery and Further Studies on Giant Viruses at the IHU Mediterranee Infection That Modified the Perception of the Virosphere , in: Viruses 11 (4), March / April 2019, pii: E312, doi: 10.3390 / v11040312 , PMC 6520786 (free full text), PMID 30935049
- ↑ a b c d e f g h i j k l m n o p q r s t u v Didier Raoult, Anthony Levasseur, Bernard La Scola: PCR Detection of Mimivirus, in: Emerging Infectious Diseases, June 2017, Vol. 23 , No. 6, pp. 1044-1045, doi: 10.3201 / eid2306.161896 , PDF
- ↑ Hansika Chhabra: Giant viruses found in water samples from Mumbai , in: BusinessLine: Science, Bangalore, May 9, 2019
- ↑ a b c d Sailen Barik: A Family of Novel Cyclophilins, Conserved in the Mimivirus Genus of the Giant DNA Viruses , in: Computational and Structural Biotechnology Journal, Volume 16, July 2018, pp. 231-236, doi: 10.1016 / j .csbj.2018.07.001
- ↑ a b Masaharu Takemura, Tatsuya Mikami, Shingo Murono: Nearly Complete Genome Sequences of Two Mimivirus Strains Isolated from a Japanese Freshwater Pond and River Mouth, in: Genome Announc. 4 (6), November / December 2016, e01378-16, doi: 10.1128 / genomeA.01378-16 , PMC 5146454 (free full text), PMID 27932662 . According to the authors, the two virus lines are closely related to Mimivirus Bombay and therefore also belong to the species APMV.
- ↑ Rafael K Campos, Paulo V Boratto, Felipe L Assis, Eric RGR Aguiar, Lorena CF Silva, Jonas D Albarnaz, Fabio P Dornas, Giliane S Trindade, Paulo P Ferreira, João T Marques, Catherine Robert, Didier Raoult, Erna G Kroon , Bernard La Scola, Jônatas S Abrahão: Samba virus: a novel mimivirus from a giant rain forest, the Brazilian Amazon , in: Virology Journal 2014 11:95, doi: 10.1186 / 1743-422X-11-95
- ↑ Jason R. Schrad, Jônatas S. Abrahão, Juliana R. Cortines and Kristin N. Parent: Structural and Proteomic Characterization of the Initiation of Giant Virus Infection , in: Cell, May 8, 2020, doi: 10.1016 / j.cell. 2020.04.032
- ↑ Jason R. Schrad, Jônatas S. Abrahão, Juliana R. Cortines, Kristin N. Parent: Boiling Acid Mimics Intracellular Giant Virus Genome Release , on: bioRxiv of September 20, 2019, doi: 10.1101 / 777854 (preprint)
- ↑ Mysterious Giant Viruses: Gargantuan in Size and Complexity , on: SciTechDaily, May 9, 2020, Source: Michigan State University
- ↑ a b c Felipe L. Assis, Leena Bajrai, Jonatas S. Abrahao, Erna G. Kroon, Fabio P. Dornas, Kétyllen R. Andrade, Paulo VM Boratto, Mariana R. Pilotto, Catherine Robert, Samia Benamar, Bernard La Scola , Philippe Colson: Pan-Genome Analysis of Brazilian Lineage A Amoebal Mimiviruses , in: Viruses 7 (7), 2015, pp. 3483-3499, doi: 10.3390 / v7072782 . According to the authors, the candidates dealt with here represent a family group, although the other sources indicate that Samba and Kroon viruses belong to the species APMV. This then applies to the other two.
- ↑ Paulo Victor Miranda Boratto, Fábio Pio Dornas, Lorena Christine Ferreira da Silva, Rodrigo Araújo Lima Rodrigues, Graziel Pereira Oliveira, Juliana Reis Cortines, Betânia Paiva Drumond, Jônatas Santos Abrahão: Analyzes of the Kroon Virus Major Capsid Gene and Its Transcript Highlight a Distinct Pattern of Gene Evolution and Splicing among Mimiviruses , in: J Virol. 92 (2), January 15, 2018, e01782-17, doi: 10.1128 / JVI.01782-17 , PMC 5752926 (free full text), PMID 29118120
- ↑ NCBI: Saudi moumouvirus
- ↑ Leena H. Bajrai, Felipe L. de Assis, Esam I. Azhar, Priscilla Jardot, Catherine Robert, Jônatas Abrahão, Didier Raoult, Bernard La Scola: Saudi Moumouvirus, the First Group B Mimivirus Isolated from Asia , in: Front. Microbiol., December 20, 2016, doi: 10.3389 / fmicb.2016.02029
- ↑ NCBI: Acanthamoeba castellanii mamavirus (species)
- ↑ a b c d Tan Yeh Fong, Chai Ying Lim, Chun Wie Chong, Patricia Kim Chooi Lim, Ivan KS Yap, Pooi Pooi Leong, Kenny Voon: Isolation and Quantification of Mimivirus-Like and Marseillevirus-Like Viruses from Soil Samples in An Aboriginal (Orang asli) Village in Peninsular Malaysia , in: Intervirology 61 (2), pp. 1-4, August 2018, doi: 10.1159 / 000491602 , Medscape , PDF , Fig. 2 - SR1, SR4 and SR9 are closely related to one another , but are much closer to the other candidates in line A than the Saudi moumouvirus (line B). The assignment to line A is therefore likely.
- ↑ a b Julien Guglielmini, Anthony Woo, Mart Krupovic, Patrick Forterre, Morgan Gaia: Diversification of giant and large eukaryotic dsDNA viruses predated the origin of modern eukaryotes , on: bioRxiv of October 29, 2018 (preprint), doi: 10.1101 / 455816
- ↑ a b Julien Guglielmini, Anthony C. Woo, Mart Krupovic, Patrick Forterre, Morgan Gaia: [1] , in: PNAS 116 (39), 10./24. September 2019, pp. 19585-19592, doi: 10.1073 / pnas.1912006116 , PMID 31506349 , Fig. 2
- ↑ NCBI: Hirudovirus strain sangsue
- ↑ Mondher Boughalmi, Isabelle Pagnier, Sarah Aherfi, Philippe Colson, Didier Raoult, Bernard La Scola: First Isolation of a Giant Virus from Wild Hirudo medicinalis Leech: Mimiviridae isolation in Hirudo medicinalis , in: Viruses 2013, 5, pp. 2920-2930 , doi: 10.3390 / v5122920
- ↑ NCBI: Niemeyer virus (species)
- ↑ Paulo VM Boratto, Thalita S. Arantes, Lorena CF Silva, Felipe L. Assis, Erna G. Kroon, Bernard La Scola, Jônatas S. Abrahão: Niemeyer Virus: A New Mimivirus Group A Isolate Harboring a Set of Duplicated Aminoacyl-tRNA Synthetase Genes , in: Front Microbiol. 2015; 6: 1256. doi: 10.3389 / fmicb.2015.01256 , PMC 4639698 (free full text), PMID 26635738
- ↑ NCBI: Mimivirus battle6 (Species)
- ↑ NCBI: Mimivirus battle7 (Species)
- ↑ NCBI: Mimivirus battle19 (Species)
- ↑ NCBI: Mimivirus battle27 (Species)
- ↑ NCBI: Mimivirus battle57 (Species)
- ↑ NCBI: Mimivirus battle66 (Species)
- ↑ NCBI: Mimivirus battle83 (Species)
- ↑ NCBI: Mimivirus battle86 (Species)
- ↑ a b c d e Anirvan Chatterjee, Thomas Sicherheitsitz-Pontén, Rajesh Yadav, Kiran Kondabagil: Isolation and complete genome sequencing of Mimivirus bombay, a Giant Virus in sewage of Mumbai, India , in: Genomics Data 9 (C), May 2016 , doi: 10.1016 / j.gdata.2016.05.013 , Fig. 2
- ↑ NCBI: Mimivirus Cher (Species)
- ↑ NCBI: Mimivirus dakar4 (Species)
- ^ NCBI: Mimivirus fauteuil
- ↑ a b c d e f Christelle Desnues, Bernard La Scola, Natalya Yutin, Ghislain Fournous et al. : Provirophages and transpovirons as the diverse mobilome of giant viruses, in: PNAS 109 (44), October 30, 2012, pp. 18078–18083, doi: 10.1073 / pnas.1208835109
- ^ NCBI: Fauteuil virus FD
- ↑ NCBI: Mimivirus huitre A06 (Species)
- ↑ NCBI: Mimivirus lactour (Species)
- ↑ NCBI: Lactours virus LT2 (species)
- ↑ NCBI: Mimivirus lentille (Species)
- ↑ NCBI: Lentille virus CL (species)
- ↑ NCBI: Mimivirus longchamps (species)
- ↑ NCBI: Longchamps virus FPL (species)
- ↑ NCBI: Mimivirus marais (species)
- ↑ NCBI: Mimivirus montadette2 (Species)
- ↑ NCBI: Mimivirus pointerouge1 (Species)
- ↑ NCBI: Pointerouge virus 1 (species)
- ↑ NCBI: Mimivirus pointerouge1 (Species)
- ↑ NCBI: Pointerouge virus 1 (species)
- ↑ NCBI: Mimivirus SR1 (Species)
- ↑ NCBI: Mimivirus SR4 (Species)
- ↑ NCBI: Mimivirus SR9 (Species)
- ↑ NCBI: Mimivirus T2 (species)
- ↑ NCBI: Mimivirus T3 (species)
- ↑ NCBI: Mimivirus terra2 (species)
- ↑ NCBI: Mimivirus univirus (Species)
- ↑ Jônatas Abrahão, Lorena Silva, Ludmila Santos Silva, Jacques Yaacoub Bou Khalil, Rodrigo Rodrigues, Thalita Arantes, Felipe Assis, Paulo Boratto, Miguel Andrade, Erna Geessien Kroon, Bergmann Ribeiro, Ivan Bergier, Herve Seligmann, Eric Ghigo, Philippe Colson, Anthony Levasseur, Guido Kroemer, Didier Raoult, Bernard La Scola: Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere . In: Nature Communications . 9, No. 1, February 27, 2018. doi : 10.1038 / s41467-018-03168-1 .

