ADAM10: Difference between revisions
m Open access bot: doi added to citation with #oabot. |
Added the cs1 style template to denote Vancouver ("vanc") citation style, because references contain "vauthors" attribute to specify the list of authors. Added doi-access. | Use this bot. | #UCB_Other |
||
| (18 intermediate revisions by 12 users not shown) | |||
| Line 1: | Line 1: | ||
{{Short description|Protein-coding gene in the species Homo sapiens}} |
|||
{{cs1 config|name-list-style=vanc|display-authors=6}} |
|||
{{Infobox_gene}} |
{{Infobox_gene}} |
||
{{enzyme |
{{infobox enzyme |
||
| Name = ADAM10 endopeptidase |
| Name = ADAM10 endopeptidase |
||
| EC_number = 3.4.24.81 |
| EC_number = 3.4.24.81 |
||
| CAS_number = 193099-09-1 |
| CAS_number = 193099-09-1 |
||
| GO_code = |
|||
| IUBMB_EC_number = 3/4/24/81 |
|||
| |
| image = |
||
| |
| width = |
||
| |
| caption = |
||
| caption = |
|||
}} |
}} |
||
'''A Disintegrin and metalloproteinase domain-containing protein 10''', also known as '''ADAM10''' or '''CDw156''' or '''CD156c''' is a [[protein]] that in humans is encoded by the ''ADAM10'' [[gene]].<ref name="entrez">{{cite web | title = Entrez Gene: ADAM10 ADAM metallopeptidase domain 10| url = https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=102}}</ref> |
'''A Disintegrin and metalloproteinase domain-containing protein 10''', also known as '''ADAM10''' or '''CDw156''' or '''CD156c''' is a [[protein]] that in humans is encoded by the ''ADAM10'' [[gene]].<ref name="entrez">{{cite web | title = Entrez Gene: ADAM10 ADAM metallopeptidase domain 10| url = https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=102}}</ref> |
||
| Line 20: | Line 21: | ||
ADAM10 (EC#: 3.4.24.81) is a [[sheddase]], and has a broad specificity for peptide hydrolysis reactions.<ref name="Brenda Database">{{cite web | title = Entry of ADAM10 endopeptidase (EC-Number 3.4.24.81 )| url = http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.4.24.81}}</ref> |
ADAM10 (EC#: 3.4.24.81) is a [[sheddase]], and has a broad specificity for peptide hydrolysis reactions.<ref name="Brenda Database">{{cite web | title = Entry of ADAM10 endopeptidase (EC-Number 3.4.24.81 )| url = http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.4.24.81}}</ref> |
||
ADAM10 cleaves [[eph receptor|ephrin]], within the ephrin/eph complex, formed between two cell surfaces. When ephrin is freed from the opposing cell, the entire ephrin/eph complex is endocytosed. This shedding in trans had not been previously shown, but may well be involved in other shedding events.<ref name="pmid16239146">{{cite journal | vauthors = Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E, Blobel CP, Himanen JP, Lackmann M, Nikolov DB |
ADAM10 cleaves [[eph receptor|ephrin]], within the ephrin/eph complex, formed between two cell surfaces. When ephrin is freed from the opposing cell, the entire ephrin/eph complex is endocytosed. This shedding in trans had not been previously shown, but may well be involved in other shedding events.<ref name="pmid16239146">{{cite journal | vauthors = Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E, Blobel CP, Himanen JP, Lackmann M, Nikolov DB | title = Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans | journal = Cell | volume = 123 | issue = 2 | pages = 291–304 | date = October 2005 | pmid = 16239146 | doi = 10.1016/j.cell.2005.08.014 | s2cid = 7962666 | doi-access = free }}</ref> |
||
In [[neuron]]s, ADAM10 is the most important enzyme, with [[alpha secretase|α-secretase]] activity for [[proteolysis|proteolytic]] processing of the [[amyloid precursor protein]].<ref>{{cite journal | vauthors = Haass C, Kaether C, Thinakaran G, Sisodia S | title = Trafficking and proteolytic processing of APP | journal = Cold Spring Harbor Perspectives in Medicine | volume = 2 | issue = 5 | pages = a006270 | date = May 2012 | pmid = 22553493 | pmc = 3331683 | doi = 10.1101/cshperspect.a006270 }}</ref> |
In [[neuron]]s, ADAM10 is the most important enzyme, with [[alpha secretase|α-secretase]] activity for [[proteolysis|proteolytic]] processing of the [[amyloid precursor protein]].<ref>{{cite journal | vauthors = Haass C, Kaether C, Thinakaran G, Sisodia S | title = Trafficking and proteolytic processing of APP | journal = Cold Spring Harbor Perspectives in Medicine | volume = 2 | issue = 5 | pages = a006270 | date = May 2012 | pmid = 22553493 | pmc = 3331683 | doi = 10.1101/cshperspect.a006270 }}</ref> ADAM10, along with [[ADAM17]], cleaves the ectodomain of the triggering receptor expressed on myeloid cells 2 ([[TREM2]]), to produce soluble TREM2 (sTREM2), which has been proposed as a CSF and sera biomarker of neurodegeneration.<ref>{{Cite journal|last1=Yang|first1=Jiaolong|last2=Fu|first2=Zhihui|last3=Zhang|first3=Xingyu|last4=Xiong|first4=Min|last5=Meng|first5=Lanxia|last6=Zhang|first6=Zhentao|date=2020-07-07|title=TREM2 ectodomain and its soluble form in Alzheimer's disease|journal=Journal of Neuroinflammation|volume=17|issue=1|pages=204|doi=10.1186/s12974-020-01878-2|issn=1742-2094|pmc=7341574|pmid=32635934 |doi-access=free }}</ref> |
||
ADAM10 belongs to subfamily A, the most ancestral subfamily of ADAM proteins, which is shared by all major groups of [[animal]]s, [[choanoflagellate]]s, [[fungi]], and [[green algae]] from the class [[Mamiellophyceae]].<ref name="ada">{{cite journal |vauthors=Souza J, Lisboa A, Santos T, Andrade M, Neves V, Teles-Souza J, Jesus H, Bezerra T, Falcão V, Oliveira R, Del-Bem L |title=The evolution of ADAM gene family in eukaryotes |journal=Genomics | year = 2020 |volume=112 |issue=5 |pages=3108–3116 | doi=10.1016/j.ygeno.2020.05.010|pmid=32437852 }}</ref> |
ADAM10 belongs to subfamily A, the most ancestral subfamily of ADAM proteins, which is shared by all major groups of [[animal]]s, [[choanoflagellate]]s, [[fungi]], and [[green algae]] from the class [[Mamiellophyceae]].<ref name="ada">{{cite journal |vauthors=Souza J, Lisboa A, Santos T, Andrade M, Neves V, Teles-Souza J, Jesus H, Bezerra T, Falcão V, Oliveira R, Del-Bem L |title=The evolution of ADAM gene family in eukaryotes |journal=Genomics | year = 2020 |volume=112 |issue=5 |pages=3108–3116 | doi=10.1016/j.ygeno.2020.05.010|pmid=32437852 |s2cid=218832838 |doi-access=free }}</ref> |
||
== Structure == |
== Structure == |
||
Although no crystallographic x-ray diffraction analyses have been published that depict the entire structure of ADAM10, one domain has been studied using this technique. The |
Although no crystallographic x-ray diffraction analyses have been published that depict the entire structure of ADAM10, one domain has been studied using this technique. The disintegrin and cysteine-rich domain (shown to the right) plays an essential role in regulation of protease activity in vivo. Recent experimental evidence suggests that this region, which is distinct from the active site, may be responsible for substrate specificity of the enzyme. It is proposed that this domain binds to particular regions of the enzyme's substrate, allowing peptide bond hydrolysis to occur in well defined locations on certain substrate proteins.<ref name="pmid12460986">{{cite journal | vauthors = Smith KM, Gaultier A, Cousin H, Alfandari D, White JM, DeSimone DW | title = The cysteine-rich domain regulates ADAM protease function in vivo | journal = The Journal of Cell Biology | volume = 159 | issue = 5 | pages = 893–902 | date = December 2002 | pmid = 12460986 | pmc = 2173380 | doi = 10.1083/jcb.200206023 }}</ref> |
||
The proposed active site of ADAM10 has been identified by sequence analysis, and is identical to enzymes in the Snake Venom metalloprotein domain family. The consensus sequence for catalytically active ADAM proteins is '''HE'''XG'''H'''NLGXX'''H'''D. Structural analysis of ADAM17, which has the same active site sequence as ADAM10, suggests that the three histidines in this sequence bind a Zn<sup>2+</sup> atom, and that the glutamate is the catalytic residue.<ref name="pmid7593158">{{cite journal | vauthors = Wolfsberg TG, Primakoff P, Myles DG, White JM | title = ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions | journal = The Journal of Cell Biology | volume = 131 | issue = 2 | pages = 275–8 | date = October 1995 | pmid = 7593158 | pmc = 2199973 | doi = 10.1083/jcb.131.2.275 }}</ref> |
The proposed active site of ADAM10 has been identified by sequence analysis, and is identical to enzymes in the Snake Venom metalloprotein domain family. The consensus sequence for catalytically active ADAM proteins is '''HE'''XG'''H'''NLGXX'''H'''D. Structural analysis of ADAM17, which has the same active site sequence as ADAM10, suggests that the three histidines in this sequence bind a Zn<sup>2+</sup> atom, and that the glutamate is the catalytic residue.<ref name="pmid7593158">{{cite journal | vauthors = Wolfsberg TG, Primakoff P, Myles DG, White JM | title = ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions | journal = The Journal of Cell Biology | volume = 131 | issue = 2 | pages = 275–8 | date = October 1995 | pmid = 7593158 | pmc = 2199973 | doi = 10.1083/jcb.131.2.275 }}</ref> |
||
| Line 37: | Line 38: | ||
In zinc proteases, the key catalytic elements have been identified as a glutamate residue and a Zn<sup>2+</sup> ion coordinated to histidine residues.<ref name="pmid2197984">{{cite journal | vauthors = Lolis E, Petsko GA | title = Transition-state analogues in protein crystallography: probes of the structural source of enzyme catalysis | journal = Annual Review of Biochemistry | volume = 59 | pages = 597–630 | year = 1990 | pmid = 2197984 | doi = 10.1146/annurev.bi.59.070190.003121 }}</ref> |
In zinc proteases, the key catalytic elements have been identified as a glutamate residue and a Zn<sup>2+</sup> ion coordinated to histidine residues.<ref name="pmid2197984">{{cite journal | vauthors = Lolis E, Petsko GA | title = Transition-state analogues in protein crystallography: probes of the structural source of enzyme catalysis | journal = Annual Review of Biochemistry | volume = 59 | pages = 597–630 | year = 1990 | pmid = 2197984 | doi = 10.1146/annurev.bi.59.070190.003121 }}</ref> |
||
The proposed mechanism begins with deprotonation of a water molecule by glutamate. The resultant hydroxide initiates a |
The proposed mechanism begins with deprotonation of a water molecule by glutamate. The resultant hydroxide initiates a nucleophilic attack on a carbonyl carbon on the peptide backbone, producing a tetrahedral intermediate. This step is facilitated by electron withdrawal from oxygen by Zn<sup>2+</sup> and by zinc's subsequent stabilization of the negative charge on the oxygen atom in the intermediate state. As electrons move down from the oxygen atom to re-form the double bond, the tetrahedral intermediate collapses to products with protonation of -NH by the glutamate residue.<ref name="pmid2197984"/> |
||
<!-- Deleted image removed: [[Image:Proposed ADAM10 mechanism.JPG|650px|border|thumb|center|Proposed Mechanism of ADAM10 Peptide Hydrolysis]] --> |
<!-- Deleted image removed: [[Image:Proposed ADAM10 mechanism.JPG|650px|border|thumb|center|Proposed Mechanism of ADAM10 Peptide Hydrolysis]] --> |
||
| Line 44: | Line 45: | ||
=== Brain diseases === |
=== Brain diseases === |
||
ADAM10 plays a key role in the modulation of the molecular mechanisms responsible for dendritic spine formation, maturation and stabilization and in the regulation of the molecular organization of the glutamatergic synapse. Consequently, an alteration of ADAM10 activity is strictly correlated to the onset of different types of synaptopathies, ranging from neurodevelopmental disorders, i.e. autism spectrum disorders, to neurodegenerative diseases, i.e. |
ADAM10 plays a key role in the modulation of the molecular mechanisms responsible for dendritic spine formation, maturation and stabilization and in the regulation of the molecular organization of the glutamatergic synapse. Consequently, an alteration of ADAM10 activity is strictly correlated to the onset of different types of synaptopathies, ranging from neurodevelopmental disorders, i.e. autism spectrum disorders, to neurodegenerative diseases, i.e. Alzheimer's Disease.<ref name="MarcelloBorroni2017">{{cite journal | vauthors = Marcello E, Borroni B, Pelucchi S, Gardoni F, Di Luca M | title = ADAM10 as a therapeutic target for brain diseases: from developmental disorders to Alzheimer's disease | journal = Expert Opinion on Therapeutic Targets | volume = 21 | issue = 11 | pages = 1017–1026 | date = November 2017 | pmid = 28960088 | doi = 10.1080/14728222.2017.1386176 | s2cid = 46800368 }}</ref> |
||
=== Interaction with the malaria parasite === |
=== Interaction with the malaria parasite === |
||
| Line 52: | Line 53: | ||
== Breast cancer == |
== Breast cancer == |
||
In combination with low doses of [[herceptin]], selective ADAM10 inhibitors decrease proliferation in [[HER2/neu|HER2]] over-expressing cell lines while inhibitors, that do not inhibit ADAM10, have no impact. These results are consistent with ADAM10 being a major determinant of HER2 shedding, the inhibition of which, may provide a novel therapeutic approach for treating breast cancer and a variety of other cancers with active HER2 signaling.<ref name="pmid16627989">{{cite journal | vauthors = Liu PC, Liu X, Li Y, Covington M, Wynn R, Huber R, Hillman M, Yang G, Ellis D, Marando C, Katiyar K, Bradley J, Abremski K, Stow M, Rupar M, Zhuo J, Li YL, Lin Q, Burns D, Xu M, Zhang C, Qian DQ, He C, Sharief V, Weng L, Agrios C, Shi E, Metcalf B, Newton R, Friedman S, Yao W, Scherle P, Hollis G, Burn TC |
In combination with low doses of [[herceptin]], selective ADAM10 inhibitors decrease proliferation in [[HER2/neu|HER2]] over-expressing cell lines while inhibitors, that do not inhibit ADAM10, have no impact. These results are consistent with ADAM10 being a major determinant of HER2 shedding, the inhibition of which, may provide a novel therapeutic approach for treating breast cancer and a variety of other cancers with active HER2 signaling.<ref name="pmid16627989">{{cite journal | vauthors = Liu PC, Liu X, Li Y, Covington M, Wynn R, Huber R, Hillman M, Yang G, Ellis D, Marando C, Katiyar K, Bradley J, Abremski K, Stow M, Rupar M, Zhuo J, Li YL, Lin Q, Burns D, Xu M, Zhang C, Qian DQ, He C, Sharief V, Weng L, Agrios C, Shi E, Metcalf B, Newton R, Friedman S, Yao W, Scherle P, Hollis G, Burn TC | title = Identification of ADAM10 as a major source of HER2 ectodomain sheddase activity in HER2 overexpressing breast cancer cells | journal = Cancer Biology & Therapy | volume = 5 | issue = 6 | pages = 657–64 | date = June 2006 | pmid = 16627989 | doi = 10.4161/cbt.5.6.2708 | s2cid = 23463401 | doi-access = free }}</ref> |
||
The presence of the product of this gene in neuronal synapses in conjunction with protein [[AP2 adaptor complex|AP2]] has been seen in increased amounts in the hippocampal neurons of [[Alzheimer's disease]] patients.<ref>{{cite journal | vauthors = Marcello E, Saraceno C, Musardo S, Vara H, de la Fuente AG, Pelucchi S, Di Marino D, Borroni B, Tramontano A, Pérez-Otaño I, Padovani A, Giustetto M, Gardoni F, Di Luca M |
The presence of the product of this gene in neuronal synapses in conjunction with protein [[AP2 adaptor complex|AP2]] has been seen in increased amounts in the hippocampal neurons of [[Alzheimer's disease]] patients.<ref>{{cite journal | vauthors = Marcello E, Saraceno C, Musardo S, Vara H, de la Fuente AG, Pelucchi S, Di Marino D, Borroni B, Tramontano A, Pérez-Otaño I, Padovani A, Giustetto M, Gardoni F, Di Luca M | title = Endocytosis of synaptic ADAM10 in neuronal plasticity and Alzheimer's disease | journal = The Journal of Clinical Investigation | volume = 123 | issue = 6 | pages = 2523–38 | date = June 2013 | pmid = 23676497 | pmc = 3668814 | doi = 10.1172/JCI65401 | url = https://hal-riip.archives-ouvertes.fr/pasteur-01054137/document }}</ref> |
||
== See also == |
== See also == |
||
| Line 69: | Line 70: | ||
* {{cite journal | vauthors = O'Bryan JP, Fridell YW, Koski R, Varnum B, Liu ET | title = The transforming receptor tyrosine kinase, Axl, is post-translationally regulated by proteolytic cleavage | journal = The Journal of Biological Chemistry | volume = 270 | issue = 2 | pages = 551–7 | date = January 1995 | pmid = 7822279 | doi = 10.1074/jbc.270.2.551 | s2cid = 46190313 | doi-access = free }} |
* {{cite journal | vauthors = O'Bryan JP, Fridell YW, Koski R, Varnum B, Liu ET | title = The transforming receptor tyrosine kinase, Axl, is post-translationally regulated by proteolytic cleavage | journal = The Journal of Biological Chemistry | volume = 270 | issue = 2 | pages = 551–7 | date = January 1995 | pmid = 7822279 | doi = 10.1074/jbc.270.2.551 | s2cid = 46190313 | doi-access = free }} |
||
* {{cite journal | vauthors = Howard L, Lu X, Mitchell S, Griffiths S, Glynn P | title = Molecular cloning of MADM: a catalytically active mammalian disintegrin-metalloprotease expressed in various cell types | journal = The Biochemical Journal | volume = 317 ( Pt 1) | issue = 1 | pages = 45–50 | date = July 1996 | pmid = 8694785 | pmc = 1217484 | doi = 10.1042/bj3170045 }} |
* {{cite journal | vauthors = Howard L, Lu X, Mitchell S, Griffiths S, Glynn P | title = Molecular cloning of MADM: a catalytically active mammalian disintegrin-metalloprotease expressed in various cell types | journal = The Biochemical Journal | volume = 317 ( Pt 1) | issue = 1 | pages = 45–50 | date = July 1996 | pmid = 8694785 | pmc = 1217484 | doi = 10.1042/bj3170045 }} |
||
* {{cite journal | vauthors = McKie N, Edwards T, Dallas DJ, Houghton A, Stringer B, Graham R, Russell G, Croucher PI |
* {{cite journal | vauthors = McKie N, Edwards T, Dallas DJ, Houghton A, Stringer B, Graham R, Russell G, Croucher PI | title = Expression of members of a novel membrane linked metalloproteinase family (ADAM) in human articular chondrocytes | journal = Biochemical and Biophysical Research Communications | volume = 230 | issue = 2 | pages = 335–9 | date = January 1997 | pmid = 9016778 | doi = 10.1006/bbrc.1996.5957 }} |
||
* {{cite journal | vauthors = Rosendahl MS, Ko SC, Long DL, Brewer MT, Rosenzweig B, Hedl E, Anderson L, Pyle SM, Moreland J, Meyers MA, Kohno T, Lyons D, Lichenstein HS |
* {{cite journal | vauthors = Rosendahl MS, Ko SC, Long DL, Brewer MT, Rosenzweig B, Hedl E, Anderson L, Pyle SM, Moreland J, Meyers MA, Kohno T, Lyons D, Lichenstein HS | title = Identification and characterization of a pro-tumor necrosis factor-alpha-processing enzyme from the ADAM family of zinc metalloproteases | journal = The Journal of Biological Chemistry | volume = 272 | issue = 39 | pages = 24588–93 | date = September 1997 | pmid = 9305925 | doi = 10.1074/jbc.272.39.24588 | s2cid = 21399815 | doi-access = free }} |
||
* {{cite journal | vauthors = Yamazaki K, Mizui Y, Tanaka I | title = Radiation hybrid mapping of human ADAM10 gene to chromosome 15 | journal = Genomics | volume = 45 | issue = 2 | pages = 457–9 | date = October 1997 | pmid = 9344679 | doi = 10.1006/geno.1997.4910 }} |
* {{cite journal | vauthors = Yamazaki K, Mizui Y, Tanaka I | title = Radiation hybrid mapping of human ADAM10 gene to chromosome 15 | journal = Genomics | volume = 45 | issue = 2 | pages = 457–9 | date = October 1997 | pmid = 9344679 | doi = 10.1006/geno.1997.4910 }} |
||
* {{cite journal | vauthors = Yamazaki K, Mizui Y, Sagane K, Tanaka I | title = Assignment of a disintegrin and metalloproteinase domain 10 (Adam10) gene to mouse chromosome 9 | journal = Genomics | volume = 46 | issue = 3 | pages = 528–9 | date = December 1997 | pmid = 9441766 | doi = 10.1006/geno.1997.5043 }} |
* {{cite journal | vauthors = Yamazaki K, Mizui Y, Sagane K, Tanaka I | title = Assignment of a disintegrin and metalloproteinase domain 10 (Adam10) gene to mouse chromosome 9 | journal = Genomics | volume = 46 | issue = 3 | pages = 528–9 | date = December 1997 | pmid = 9441766 | doi = 10.1006/geno.1997.5043 }} |
||
* {{cite journal | vauthors = Yavari R, Adida C, Bray-Ward P, Brines M, Xu T | title = Human metalloprotease-disintegrin Kuzbanian regulates sympathoadrenal cell fate in development and neoplasia | journal = Human Molecular Genetics | volume = 7 | issue = 7 | pages = 1161–7 | date = July 1998 | pmid = 9618175 | doi = 10.1093/hmg/7.7.1161 | doi-access = free }} |
* {{cite journal | vauthors = Yavari R, Adida C, Bray-Ward P, Brines M, Xu T | title = Human metalloprotease-disintegrin Kuzbanian regulates sympathoadrenal cell fate in development and neoplasia | journal = Human Molecular Genetics | volume = 7 | issue = 7 | pages = 1161–7 | date = July 1998 | pmid = 9618175 | doi = 10.1093/hmg/7.7.1161 | doi-access = free }} |
||
* {{cite journal | vauthors = Dallas DJ, Genever PG, Patton AJ, Millichip MI, McKie N, Skerry TM | title = Localization of ADAM10 and Notch receptors in bone | journal = Bone | volume = 25 | issue = 1 | pages = 9–15 | date = July 1999 | pmid = 10423016 | doi = 10.1016/S8756-3282(99)00099-X }} |
* {{cite journal | vauthors = Dallas DJ, Genever PG, Patton AJ, Millichip MI, McKie N, Skerry TM | title = Localization of ADAM10 and Notch receptors in bone | journal = Bone | volume = 25 | issue = 1 | pages = 9–15 | date = July 1999 | pmid = 10423016 | doi = 10.1016/S8756-3282(99)00099-X }} |
||
* {{cite journal | vauthors = Dias Neto E, Correa RG, Verjovski-Almeida S, Briones MR, Nagai MA, da Silva W, Zago MA, Bordin S, Costa FF, Goldman GH, Carvalho AF, Matsukuma A, Baia GS, Simpson DH, Brunstein A, de Oliveira PS, Bucher P, Jongeneel CV, O'Hare MJ, Soares F, Brentani RR, Reis LF, de Souza SJ, Simpson AJ |
* {{cite journal | vauthors = Dias Neto E, Correa RG, Verjovski-Almeida S, Briones MR, Nagai MA, da Silva W, Zago MA, Bordin S, Costa FF, Goldman GH, Carvalho AF, Matsukuma A, Baia GS, Simpson DH, Brunstein A, de Oliveira PS, Bucher P, Jongeneel CV, O'Hare MJ, Soares F, Brentani RR, Reis LF, de Souza SJ, Simpson AJ | title = Shotgun sequencing of the human transcriptome with ORF expressed sequence tags | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 97 | issue = 7 | pages = 3491–6 | date = March 2000 | pmid = 10737800 | pmc = 16267 | doi = 10.1073/pnas.97.7.3491 | bibcode = 2000PNAS...97.3491D | doi-access = free }} |
||
* {{cite journal | vauthors = Hattori M, Osterfield M, Flanagan JG | title = Regulated cleavage of a contact-mediated axon repellent | journal = Science | volume = 289 | issue = 5483 | pages = 1360–5 | date = August 2000 | pmid = 10958785 | doi = 10.1126/science.289.5483.1360 | bibcode = 2000Sci...289.1360H }} |
* {{cite journal | vauthors = Hattori M, Osterfield M, Flanagan JG | title = Regulated cleavage of a contact-mediated axon repellent | journal = Science | volume = 289 | issue = 5483 | pages = 1360–5 | date = August 2000 | pmid = 10958785 | doi = 10.1126/science.289.5483.1360 | bibcode = 2000Sci...289.1360H }} |
||
* {{cite journal | vauthors = Vincent B, Paitel E, Saftig P, Frobert Y, Hartmann D, De Strooper B, Grassi J, Lopez-Perez E, Checler F |
* {{cite journal | vauthors = Vincent B, Paitel E, Saftig P, Frobert Y, Hartmann D, De Strooper B, Grassi J, Lopez-Perez E, Checler F | title = The disintegrins ADAM10 and TACE contribute to the constitutive and phorbol ester-regulated normal cleavage of the cellular prion protein | journal = The Journal of Biological Chemistry | volume = 276 | issue = 41 | pages = 37743–6 | date = October 2001 | pmid = 11477090 | doi = 10.1074/jbc.M105677200 | doi-access = free }} |
||
* {{cite journal | vauthors = Chubinskaya S, Mikhail R, Deutsch A, Tindal MH | title = ADAM-10 protein is present in human articular cartilage primarily in the membrane-bound form and is upregulated in osteoarthritis and in response to IL-1alpha in bovine nasal cartilage | journal = The Journal of Histochemistry and Cytochemistry | volume = 49 | issue = 9 | pages = 1165–76 | date = September 2001 | pmid = 11511685 | doi = 10.1177/002215540104900910 | s2cid = 6879742 | doi-access = free }} |
* {{cite journal | vauthors = Chubinskaya S, Mikhail R, Deutsch A, Tindal MH | title = ADAM-10 protein is present in human articular cartilage primarily in the membrane-bound form and is upregulated in osteoarthritis and in response to IL-1alpha in bovine nasal cartilage | journal = The Journal of Histochemistry and Cytochemistry | volume = 49 | issue = 9 | pages = 1165–76 | date = September 2001 | pmid = 11511685 | doi = 10.1177/002215540104900910 | s2cid = 6879742 | doi-access = free }} |
||
* {{cite journal | vauthors = Lemjabbar H, Basbaum C | title = Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells | journal = Nature Medicine | volume = 8 | issue = 1 | pages = 41–6 | date = January 2002 | pmid = 11786905 | doi = 10.1038/nm0102-41 | s2cid = 7135441 }} |
* {{cite journal | vauthors = Lemjabbar H, Basbaum C | title = Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells | journal = Nature Medicine | volume = 8 | issue = 1 | pages = 41–6 | date = January 2002 | pmid = 11786905 | doi = 10.1038/nm0102-41 | s2cid = 7135441 }} |
||
* {{cite journal | vauthors = Healy EF, Romano P, Mejia M, Lindfors G | title = Acetylenic inhibitors of ADAM10 and ADAM17: in silico analysis of potency and selectivity | journal = Journal of Molecular Graphics & Modelling | volume = 29 | issue = 3 | pages = 436–42 | date = November 2010 | pmid = 20863729 | doi = 10.1016/j.jmgm.2010.08.006 }} |
* {{cite journal | vauthors = Healy EF, Romano P, Mejia M, Lindfors G | title = Acetylenic inhibitors of ADAM10 and ADAM17: in silico analysis of potency and selectivity | journal = Journal of Molecular Graphics & Modelling | volume = 29 | issue = 3 | pages = 436–42 | date = November 2010 | pmid = 20863729 | doi = 10.1016/j.jmgm.2010.08.006 }} |
||
* {{cite journal | vauthors = Arndt M, Lendeckel U, Röcken C, Nepple K, Wolke C, Spiess A, Huth C, Ansorge S, Klein HU, Goette A |
* {{cite journal | vauthors = Arndt M, Lendeckel U, Röcken C, Nepple K, Wolke C, Spiess A, Huth C, Ansorge S, Klein HU, Goette A | title = Altered expression of ADAMs (A Disintegrin And Metalloproteinase) in fibrillating human atria | journal = Circulation | volume = 105 | issue = 6 | pages = 720–5 | date = February 2002 | pmid = 11839628 | doi = 10.1161/hc0602.103639 | s2cid = 10279346 | doi-access = free }} |
||
* {{cite journal | vauthors = Colciaghi F, Borroni B, Pastorino L, Marcello E, Zimmermann M, Cattabeni F, Padovani A, Di Luca M |
* {{cite journal | vauthors = Colciaghi F, Borroni B, Pastorino L, Marcello E, Zimmermann M, Cattabeni F, Padovani A, Di Luca M | title = [alpha]-Secretase ADAM10 as well as [alpha]APPs is reduced in platelets and CSF of Alzheimer disease patients | journal = Molecular Medicine | volume = 8 | issue = 2 | pages = 67–74 | date = February 2002 | pmid = 12080182 | pmc = 2039975 | doi = 10.1007/BF03402076 }} |
||
* {{cite journal | vauthors = Lim R, Winteringham LN, Williams JH, McCulloch RK, Ingley E, Tiao JY, Lalonde JP, Tsai S, Tilbrook PA, Sun Y, Wu X, Morris SW, Klinken SP |
* {{cite journal | vauthors = Lim R, Winteringham LN, Williams JH, McCulloch RK, Ingley E, Tiao JY, Lalonde JP, Tsai S, Tilbrook PA, Sun Y, Wu X, Morris SW, Klinken SP | title = MADM, a novel adaptor protein that mediates phosphorylation of the 14-3-3 binding site of myeloid leukemia factor 1 | journal = The Journal of Biological Chemistry | volume = 277 | issue = 43 | pages = 40997–1008 | date = October 2002 | pmid = 12176995 | doi = 10.1074/jbc.M206041200 | s2cid = 40077371 | url = http://researchrepository.murdoch.edu.au/id/eprint/39572/1/MADM-novel-adaptor-protein.pdf | doi-access = free }} |
||
* {{cite journal | vauthors = Gatta LB, Albertini A, Ravid R, Finazzi D | title = Levels of beta-secretase BACE and alpha-secretase ADAM10 mRNAs in Alzheimer hippocampus | journal = NeuroReport | volume = 13 | issue = 16 | pages = 2031–3 | date = November 2002 | pmid = 12438920 | doi = 10.1097/00001756-200211150-00008 }} |
* {{cite journal | vauthors = Gatta LB, Albertini A, Ravid R, Finazzi D | title = Levels of beta-secretase BACE and alpha-secretase ADAM10 mRNAs in Alzheimer hippocampus | journal = NeuroReport | volume = 13 | issue = 16 | pages = 2031–3 | date = November 2002 | pmid = 12438920 | doi = 10.1097/00001756-200211150-00008 }} |
||
* {{cite journal | vauthors = Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, Joumaa S, Zentgraf H, Fogel M, Altevogt DP |
* {{cite journal | vauthors = Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, Joumaa S, Zentgraf H, Fogel M, Altevogt DP | title = ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles | journal = FASEB Journal | volume = 17 | issue = 2 | pages = 292–4 | date = February 2003 | pmid = 12475894 | doi = 10.1096/fj.02-0430fje | doi-access = free | s2cid = 21521110 }} |
||
{{refend}} |
{{refend}} |
||
| Line 101: | Line 102: | ||
{{DEFAULTSORT:Adam10}} |
{{DEFAULTSORT:Adam10}} |
||
[[Category: |
[[Category:Proteases]] |
||
[[Category:Clusters of differentiation]] |
[[Category:Clusters of differentiation]] |
||
[[Category:Human proteins]] |
[[Category:Human proteins]] |
||
Latest revision as of 02:52, 9 February 2024
| ADAM10 endopeptidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 3.4.24.81 | ||||||||
| CAS no. | 193099-09-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
A Disintegrin and metalloproteinase domain-containing protein 10, also known as ADAM10 or CDw156 or CD156c is a protein that in humans is encoded by the ADAM10 gene.[5]
Function[edit]
Members of the ADAM family are cell surface proteins with a unique structure, possessing both potential adhesion and protease domains. Sheddase, a generic name for the ADAM metallopeptidase, functions primarily to cleave membrane proteins at the cellular surface. Once cleaved, the sheddases release soluble ectodomains with an altered location and function.[6][7][8]
Although a single sheddase may “shed” a variety of substances, multiple sheddases can cleave the same substrate resulting in different consequences. This gene encodes an ADAM family member that cleaves many proteins including TNF-alpha and E-cadherin.[5]
ADAM10 (EC#: 3.4.24.81) is a sheddase, and has a broad specificity for peptide hydrolysis reactions.[9]
ADAM10 cleaves ephrin, within the ephrin/eph complex, formed between two cell surfaces. When ephrin is freed from the opposing cell, the entire ephrin/eph complex is endocytosed. This shedding in trans had not been previously shown, but may well be involved in other shedding events.[10]
In neurons, ADAM10 is the most important enzyme, with α-secretase activity for proteolytic processing of the amyloid precursor protein.[11] ADAM10, along with ADAM17, cleaves the ectodomain of the triggering receptor expressed on myeloid cells 2 (TREM2), to produce soluble TREM2 (sTREM2), which has been proposed as a CSF and sera biomarker of neurodegeneration.[12]
ADAM10 belongs to subfamily A, the most ancestral subfamily of ADAM proteins, which is shared by all major groups of animals, choanoflagellates, fungi, and green algae from the class Mamiellophyceae.[13]
Structure[edit]
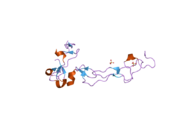
Although no crystallographic x-ray diffraction analyses have been published that depict the entire structure of ADAM10, one domain has been studied using this technique. The disintegrin and cysteine-rich domain (shown to the right) plays an essential role in regulation of protease activity in vivo. Recent experimental evidence suggests that this region, which is distinct from the active site, may be responsible for substrate specificity of the enzyme. It is proposed that this domain binds to particular regions of the enzyme's substrate, allowing peptide bond hydrolysis to occur in well defined locations on certain substrate proteins.[14]
The proposed active site of ADAM10 has been identified by sequence analysis, and is identical to enzymes in the Snake Venom metalloprotein domain family. The consensus sequence for catalytically active ADAM proteins is HEXGHNLGXXHD. Structural analysis of ADAM17, which has the same active site sequence as ADAM10, suggests that the three histidines in this sequence bind a Zn2+ atom, and that the glutamate is the catalytic residue.[15]
Catalytic Mechanism[edit]
Although the exact mechanism of ADAM10 has not been thoroughly investigated, its active site is homologous to those of well studied zinc-proteases such as carboxypeptidase A and thermolysin. Therefore, it is proposed that ADAM10 utilizes a similar mechanism as these enzymes. In zinc proteases, the key catalytic elements have been identified as a glutamate residue and a Zn2+ ion coordinated to histidine residues.[16]
The proposed mechanism begins with deprotonation of a water molecule by glutamate. The resultant hydroxide initiates a nucleophilic attack on a carbonyl carbon on the peptide backbone, producing a tetrahedral intermediate. This step is facilitated by electron withdrawal from oxygen by Zn2+ and by zinc's subsequent stabilization of the negative charge on the oxygen atom in the intermediate state. As electrons move down from the oxygen atom to re-form the double bond, the tetrahedral intermediate collapses to products with protonation of -NH by the glutamate residue.[16]
Clinical significance[edit]
Brain diseases[edit]
ADAM10 plays a key role in the modulation of the molecular mechanisms responsible for dendritic spine formation, maturation and stabilization and in the regulation of the molecular organization of the glutamatergic synapse. Consequently, an alteration of ADAM10 activity is strictly correlated to the onset of different types of synaptopathies, ranging from neurodevelopmental disorders, i.e. autism spectrum disorders, to neurodegenerative diseases, i.e. Alzheimer's Disease.[17]
Interaction with the malaria parasite[edit]
A number of different proteins on the surface of Plasmodium falciparum malaria parasites help the invaders bind to red blood cells. But once attached to host blood cells, the parasites need to shed the 'sticky' surface proteins that would otherwise interfere with entrance into the cell. The Sheddase enzyme, specifically called PfSUB2 in this example, is required for the parasites to invade cells; without it, the parasites die. The sheddase is stored in and released from cellular compartments near the tip of the parasite, according to the study. Once on the surface, the enzyme attaches to a motor that shuttles it from front to back, liberating the sticky surface proteins. With these proteins removed, the parasite gains entrance into a red blood cell. The entire invasion lasts about 30 seconds and without this ADAM metallopeptidase, malaria would be ineffective at invading the red blood cells.[18]
Breast cancer[edit]
In combination with low doses of herceptin, selective ADAM10 inhibitors decrease proliferation in HER2 over-expressing cell lines while inhibitors, that do not inhibit ADAM10, have no impact. These results are consistent with ADAM10 being a major determinant of HER2 shedding, the inhibition of which, may provide a novel therapeutic approach for treating breast cancer and a variety of other cancers with active HER2 signaling.[19]
The presence of the product of this gene in neuronal synapses in conjunction with protein AP2 has been seen in increased amounts in the hippocampal neurons of Alzheimer's disease patients.[20]
See also[edit]
References[edit]
- ^ a b c GRCh38: Ensembl release 89: ENSG00000137845 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000054693 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b "Entrez Gene: ADAM10 ADAM metallopeptidase domain 10".
- ^ Moss ML, Bartsch JW (June 2004). "Therapeutic benefits from targeting of ADAM family members". Biochemistry. 43 (23): 7227–35. doi:10.1021/bi049677f. PMID 15182168.
- ^ Nagano O, Saya H (December 2004). "Mechanism and biological significance of CD44 cleavage". Cancer Science. 95 (12): 930–5. doi:10.1111/j.1349-7006.2004.tb03179.x. PMID 15596040. S2CID 9009213.
- ^ Blobel CP (January 2005). "ADAMs: key components in EGFR signalling and development". Nature Reviews. Molecular Cell Biology. 6 (1): 32–43. doi:10.1038/nrm1548. PMID 15688065. S2CID 7011302.
- ^ "Entry of ADAM10 endopeptidase (EC-Number 3.4.24.81 )".
- ^ Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E, et al. (October 2005). "Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans". Cell. 123 (2): 291–304. doi:10.1016/j.cell.2005.08.014. PMID 16239146. S2CID 7962666.
- ^ Haass C, Kaether C, Thinakaran G, Sisodia S (May 2012). "Trafficking and proteolytic processing of APP". Cold Spring Harbor Perspectives in Medicine. 2 (5): a006270. doi:10.1101/cshperspect.a006270. PMC 3331683. PMID 22553493.
- ^ Yang J, Fu Z, Zhang X, Xiong M, Meng L, Zhang Z (2020-07-07). "TREM2 ectodomain and its soluble form in Alzheimer's disease". Journal of Neuroinflammation. 17 (1): 204. doi:10.1186/s12974-020-01878-2. ISSN 1742-2094. PMC 7341574. PMID 32635934.
- ^ Souza J, Lisboa A, Santos T, Andrade M, Neves V, Teles-Souza J, et al. (2020). "The evolution of ADAM gene family in eukaryotes". Genomics. 112 (5): 3108–3116. doi:10.1016/j.ygeno.2020.05.010. PMID 32437852. S2CID 218832838.
- ^ Smith KM, Gaultier A, Cousin H, Alfandari D, White JM, DeSimone DW (December 2002). "The cysteine-rich domain regulates ADAM protease function in vivo". The Journal of Cell Biology. 159 (5): 893–902. doi:10.1083/jcb.200206023. PMC 2173380. PMID 12460986.
- ^ Wolfsberg TG, Primakoff P, Myles DG, White JM (October 1995). "ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions". The Journal of Cell Biology. 131 (2): 275–8. doi:10.1083/jcb.131.2.275. PMC 2199973. PMID 7593158.
- ^ a b Lolis E, Petsko GA (1990). "Transition-state analogues in protein crystallography: probes of the structural source of enzyme catalysis". Annual Review of Biochemistry. 59: 597–630. doi:10.1146/annurev.bi.59.070190.003121. PMID 2197984.
- ^ Marcello E, Borroni B, Pelucchi S, Gardoni F, Di Luca M (November 2017). "ADAM10 as a therapeutic target for brain diseases: from developmental disorders to Alzheimer's disease". Expert Opinion on Therapeutic Targets. 21 (11): 1017–1026. doi:10.1080/14728222.2017.1386176. PMID 28960088. S2CID 46800368.
- ^ "'Sheddase' helps the malaria parasite invade red blood cells". Archived from the original on 2008-04-12.
- ^ Liu PC, Liu X, Li Y, Covington M, Wynn R, Huber R, et al. (June 2006). "Identification of ADAM10 as a major source of HER2 ectodomain sheddase activity in HER2 overexpressing breast cancer cells". Cancer Biology & Therapy. 5 (6): 657–64. doi:10.4161/cbt.5.6.2708. PMID 16627989. S2CID 23463401.
- ^ Marcello E, Saraceno C, Musardo S, Vara H, de la Fuente AG, Pelucchi S, et al. (June 2013). "Endocytosis of synaptic ADAM10 in neuronal plasticity and Alzheimer's disease". The Journal of Clinical Investigation. 123 (6): 2523–38. doi:10.1172/JCI65401. PMC 3668814. PMID 23676497.
Further reading[edit]
- Wolfsberg TG, Primakoff P, Myles DG, White JM (October 1995). "ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions". The Journal of Cell Biology. 131 (2): 275–8. doi:10.1083/jcb.131.2.275. PMC 2199973. PMID 7593158.
- O'Bryan JP, Fridell YW, Koski R, Varnum B, Liu ET (January 1995). "The transforming receptor tyrosine kinase, Axl, is post-translationally regulated by proteolytic cleavage". The Journal of Biological Chemistry. 270 (2): 551–7. doi:10.1074/jbc.270.2.551. PMID 7822279. S2CID 46190313.
- Howard L, Lu X, Mitchell S, Griffiths S, Glynn P (July 1996). "Molecular cloning of MADM: a catalytically active mammalian disintegrin-metalloprotease expressed in various cell types". The Biochemical Journal. 317 ( Pt 1) (1): 45–50. doi:10.1042/bj3170045. PMC 1217484. PMID 8694785.
- McKie N, Edwards T, Dallas DJ, Houghton A, Stringer B, Graham R, et al. (January 1997). "Expression of members of a novel membrane linked metalloproteinase family (ADAM) in human articular chondrocytes". Biochemical and Biophysical Research Communications. 230 (2): 335–9. doi:10.1006/bbrc.1996.5957. PMID 9016778.
- Rosendahl MS, Ko SC, Long DL, Brewer MT, Rosenzweig B, Hedl E, et al. (September 1997). "Identification and characterization of a pro-tumor necrosis factor-alpha-processing enzyme from the ADAM family of zinc metalloproteases". The Journal of Biological Chemistry. 272 (39): 24588–93. doi:10.1074/jbc.272.39.24588. PMID 9305925. S2CID 21399815.
- Yamazaki K, Mizui Y, Tanaka I (October 1997). "Radiation hybrid mapping of human ADAM10 gene to chromosome 15". Genomics. 45 (2): 457–9. doi:10.1006/geno.1997.4910. PMID 9344679.
- Yamazaki K, Mizui Y, Sagane K, Tanaka I (December 1997). "Assignment of a disintegrin and metalloproteinase domain 10 (Adam10) gene to mouse chromosome 9". Genomics. 46 (3): 528–9. doi:10.1006/geno.1997.5043. PMID 9441766.
- Yavari R, Adida C, Bray-Ward P, Brines M, Xu T (July 1998). "Human metalloprotease-disintegrin Kuzbanian regulates sympathoadrenal cell fate in development and neoplasia". Human Molecular Genetics. 7 (7): 1161–7. doi:10.1093/hmg/7.7.1161. PMID 9618175.
- Dallas DJ, Genever PG, Patton AJ, Millichip MI, McKie N, Skerry TM (July 1999). "Localization of ADAM10 and Notch receptors in bone". Bone. 25 (1): 9–15. doi:10.1016/S8756-3282(99)00099-X. PMID 10423016.
- Dias Neto E, Correa RG, Verjovski-Almeida S, Briones MR, Nagai MA, da Silva W, et al. (March 2000). "Shotgun sequencing of the human transcriptome with ORF expressed sequence tags". Proceedings of the National Academy of Sciences of the United States of America. 97 (7): 3491–6. Bibcode:2000PNAS...97.3491D. doi:10.1073/pnas.97.7.3491. PMC 16267. PMID 10737800.
- Hattori M, Osterfield M, Flanagan JG (August 2000). "Regulated cleavage of a contact-mediated axon repellent". Science. 289 (5483): 1360–5. Bibcode:2000Sci...289.1360H. doi:10.1126/science.289.5483.1360. PMID 10958785.
- Vincent B, Paitel E, Saftig P, Frobert Y, Hartmann D, De Strooper B, et al. (October 2001). "The disintegrins ADAM10 and TACE contribute to the constitutive and phorbol ester-regulated normal cleavage of the cellular prion protein". The Journal of Biological Chemistry. 276 (41): 37743–6. doi:10.1074/jbc.M105677200. PMID 11477090.
- Chubinskaya S, Mikhail R, Deutsch A, Tindal MH (September 2001). "ADAM-10 protein is present in human articular cartilage primarily in the membrane-bound form and is upregulated in osteoarthritis and in response to IL-1alpha in bovine nasal cartilage". The Journal of Histochemistry and Cytochemistry. 49 (9): 1165–76. doi:10.1177/002215540104900910. PMID 11511685. S2CID 6879742.
- Lemjabbar H, Basbaum C (January 2002). "Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells". Nature Medicine. 8 (1): 41–6. doi:10.1038/nm0102-41. PMID 11786905. S2CID 7135441.
- Healy EF, Romano P, Mejia M, Lindfors G (November 2010). "Acetylenic inhibitors of ADAM10 and ADAM17: in silico analysis of potency and selectivity". Journal of Molecular Graphics & Modelling. 29 (3): 436–42. doi:10.1016/j.jmgm.2010.08.006. PMID 20863729.
- Arndt M, Lendeckel U, Röcken C, Nepple K, Wolke C, Spiess A, et al. (February 2002). "Altered expression of ADAMs (A Disintegrin And Metalloproteinase) in fibrillating human atria". Circulation. 105 (6): 720–5. doi:10.1161/hc0602.103639. PMID 11839628. S2CID 10279346.
- Colciaghi F, Borroni B, Pastorino L, Marcello E, Zimmermann M, Cattabeni F, et al. (February 2002). "[alpha]-Secretase ADAM10 as well as [alpha]APPs is reduced in platelets and CSF of Alzheimer disease patients". Molecular Medicine. 8 (2): 67–74. doi:10.1007/BF03402076. PMC 2039975. PMID 12080182.
- Lim R, Winteringham LN, Williams JH, McCulloch RK, Ingley E, Tiao JY, et al. (October 2002). "MADM, a novel adaptor protein that mediates phosphorylation of the 14-3-3 binding site of myeloid leukemia factor 1" (PDF). The Journal of Biological Chemistry. 277 (43): 40997–1008. doi:10.1074/jbc.M206041200. PMID 12176995. S2CID 40077371.
- Gatta LB, Albertini A, Ravid R, Finazzi D (November 2002). "Levels of beta-secretase BACE and alpha-secretase ADAM10 mRNAs in Alzheimer hippocampus". NeuroReport. 13 (16): 2031–3. doi:10.1097/00001756-200211150-00008. PMID 12438920.
- Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, Joumaa S, et al. (February 2003). "ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles". FASEB Journal. 17 (2): 292–4. doi:10.1096/fj.02-0430fje. PMID 12475894. S2CID 21521110.
External links[edit]
- ADAM10 human gene location in the UCSC Genome Browser.
- ADAM10 human gene details in the UCSC Genome Browser.
- Overview of all the structural information available in the PDB for UniProt: O14672 (Disintegrin and metalloproteinase domain-containing protein 10) at the PDBe-KB.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.