Frederick Sanger
Frederick Sanger OM , CH , CBE (born August 13, 1918 in Rendcomb , Gloucestershire , † November 19, 2013 in Cambridge , Cambridgeshire ) was a British biochemist .
He was one of the few people who was honored twice with the Nobel Prize: In 1958 Sanger received the Nobel Prize in Chemistry (as the sole winner) for the elucidation of the structure of insulin and his work on protein sequencing . In 1980 he was again awarded the Nobel Prize for Chemistry (together with Paul Berg , born 1926, and Walter Gilbert , born 1932), this time for investigations into the determination of the base sequence in nucleic acids .
Life
School and study
Frederick Sanger was the second son of doctor Dr. Frederick Sanger senior (1876–1937) and Cicely Sanger (1880–1938) were born in Rendscomb. Influenced by his father and his one year older brother Theodore, Sanger developed an interest in the natural sciences at an early age . After completing school at Bryanston School and, from 1936, at St John's College in Cambridge , he originally wanted to study medicine, but then decided on biochemistry , since as a scientist, unlike in the medical profession, he could concentrate more on one topic and thus perhaps achieve more . So Sanger began studying biochemistry at the Department of Biochemistry in Cambridge.
In 1939, Sanger received his Bachelor of Arts degree . Since he came from a Quaker family, he refused military service for reasons of conscience and continued to work on his doctoral thesis during the Second World War , which he was doing in the same institute under the supervision of A. Neuberger on the metabolism of the amino acid lysine . In 1943 he received his doctorate .
Research activity
Sanger's work was supported by a grant from the Beit Memorial Fellowship for Medical Research from 1944 to 1951 . In 1951 he became an external employee of the Medical Research Council (MRC).
In the year of his PhD , Albert Chibnall succeeded Frederick Gowland Hopkins as director of biochemistry at Cambridge, and Sanger became a member of Chibnall's research group. The main interest of the group was protein chemistry, especially that of insulin . In 1945 Sanger finally developed a method for determining the amino acid sequence , with the help of which he completely determined the insulin sequence over a period of twelve years. In 1955 the sequence of the 51 amino acids arranged in a chain was published in insulin, for which Sanger was awarded the Nobel Prize in Chemistry in 1958 .
As a result, Sanger stayed in Cambridge and in 1962 became head of the protein chemistry department at the Laboratory of Molecular Biology (LMB). This institute was built in 1962 as a new laboratory complex after the Medical Research Council had set up a group in Cambridge in 1947 to “study the molecular structure of biological systems”. Although Sanger had not been particularly interested in nucleic acids until then , through discussions with scientists such as Francis Crick and Sydney Brenner, he recognized the need to determine the sequence of this other biopolymer as well. In the following years Sanger therefore devoted himself to the development of another sequencing method, which finally led to the " chain termination method" in 1975 . In 1977, DNA sequencing based on the completely deciphered genome of a bacteriophage was presented to the world for the first time. In 1980, Sanger was awarded the Nobel Prize in Chemistry for the second time for his contributions to the sequencing of nucleic acids.
In 1983 Frederick Sanger retired. Most recently, he and his wife Margaret Joan devoted themselves to his hobbies: gardening and sailing. They have three children from their marriage. Frederick Sanger died on November 19, 2013 in Cambridge.
Awards
- 1951 Corday Morgan Medal from the Royal Society of Chemistry
- 1954 Appointment as Fellow of the Royal Society (FRS)
- 1958 member of the American Academy of Arts and Sciences
- 1958 and 1980: Nobel Prize in Chemistry. In the history of the Nobel Prize, only four scientists have managed to receive this highest honor twice (besides Sanger these were Marie Curie , Linus Pauling and John Bardeen . Sanger and Bardeen are also the only people who have been awarded the Nobel Prize twice in the same discipline were).
- 1963 Awarded the Commander of the Order of the British Empire (CBE) by Queen Elizabeth II.
- 1967 member of the National Academy of Sciences
- In 1969 Sanger received the Royal Medal from the Royal Society .
- 1971, 1979 Gairdner Foundation International Award
- 1977 Copley Medal from the Royal Society
- 1979 Albert Lasker Award for Basic Medical Research
- 1981 Admission to the Order of the Companions of Honor (CH)
- In 1986, Sanger was accepted into the Order of Merit (OM) by Elizabeth II , whose regular membership is limited to just 24 people.
- In 1988 Sanger became a corresponding member of the Bavarian Academy of Sciences .
- The Sanger Center , named after Frederick Sanger, was founded in Cambridge in 1992 with the aim of sequencing and researching the human genome and the genomes of other organisms.
- In 1997, the Department of Biochemistry at Cambridge University, where Sanger had worked until 1962, was expanded. In addition to the “Hopkins Building”, there is now a second, the “Sanger Building”.
plant
Method for determining the amino acid sequence
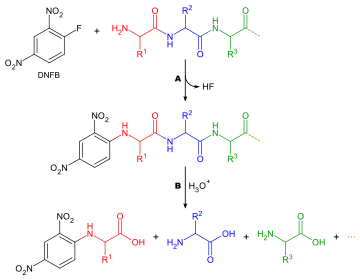
Sanger's method for determining the amino acid sequence originated during his time as a member of Albert Chibnall's research group in Cambridge. In the 1940s, protein chemistry experienced a revolution with the development of efficient chromatographic separation methods for proteins , peptides and amino acids . Sanger now wanted to determine the order of the amino acids in a protein ( amino acid sequence ). To do this, he first split the protein chain into small peptide fragments and then isolated them using the new methods. To mark and later identify the terminal amino acid, Sanger converted the fragment peptides with 1-fluoro-2,4-dinitrobenzene . The peptides derivatized at the N terminus were completely cleaved into their amino acids, the relative amounts of which were then quantified. The identity of the terminal amino acid could be determined by chromatographic analysis of the colored dinitrophenyl (DNP) derivative. In this operation, two pieces of information were obtained: 1. the identity of the first amino acid in the chain and 2. the nature of the other amino acids in the chain (although not their position). By repeating the process several times, conclusions can be drawn about the original sequence. The figure illustrates the principle of the sequencing method on a small peptide.
In twelve years of work, Sanger succeeded in determining the complete sequence of insulin. To make matters worse, insulin is a protein that consists of two polypeptide chains that are linked by disulfide bridges . The arrangement of these bridges also had to be determined. In 1955 the complete insulin sequence was published. This was the first time to prove that proteins have a unique chemical structure. Earlier hypotheses could now be finally rejected, such as the one that proteins have a defined amino acid composition, but with a random sequence, or that proteins are even aggregates of smaller, similar units. In 1958 Sanger was awarded the Nobel Prize in Chemistry for this work .
Sanger's sequencing method was later superseded by the phenyl isothiocyanate degradation developed by the Swedish biochemist Pehr Edman . The main advantage of the Edman method was that amino acids could be successively broken down and identified from the N terminus. The remaining peptide, shortened by one amino acid, could then be subjected to another degradation cycle and the sequence could be determined relatively quickly.
Method for sequencing nucleic acids
→ Main articles: DNA sequencing, Sanger dideoxy method
Sanger's method for sequencing nucleic acids originated during his time as head of the protein chemistry department at the Laboratory of Molecular Biology (LMB) in Cambridge. It arose from the need to determine the sequence of the nucleobases .
To achieve this goal, Sanger first developed a method of sequencing ribonucleic acids (RNA), which he then applied to deoxyribonucleic acid (DNA). However, this method was very slow and only allowed the determination of short sequence sections. In the following years he developed a new method that was to become the basis for today's DNA sequencing method, the so-called dideoxy method. This technique uses the following properties of DNA:
- DNA is a double-stranded molecule , not unlike a winding rope ladder. The rungs of the ladder form pairs of nucleobases ( adenine , guanine , thymine and cytosine ), of which there are only two combinations: adenine is opposite thymine, cytosine is paired with guanine.
- If the two single strands are separated, the complementary strand can be synthesized with the aid of a DNA polymerase . However, you need a small complementary piece of DNA ( primer ) that is bound to the single strand and extended by the polymerase according to the instruction of the opposite strand (template strand).
The synthesized piece of DNA has a defined starting point. The synthesis products, however, can have different lengths, the length depends on how many free nucleotides are available for synthesis or whether the polymerase accidentally falls off the template strand.

dNTP is the general abbreviation for a nucleoside triphosphate and can stand for dATP, dCTP, dGTP or dTTP. ddNTPs are the corresponding dideoxy variants of the dNTPs. The incorporation of a ddNTP leads to the termination of the polymerization reaction. The blue dots at the 5 'end of the primer represent a marking (e.g. a fluorescent group) by means of which the synthesis products can later be made visible in the gel. Alternatively, radioactively labeled nucleoside triphosphates can also be used for the polymerization reaction.
Sanger's trick now is to carry out the polymerization reaction in four separate approaches, and to ensure that each strand has the same start (which is given by the primer) and that the extension reaction - although at different points - always occurs on a certain type of base should end. To ensure this, each reaction mixture contains the four nucleotide monomers as well as the dideoxy variant of a nucleotide type. The chain extension continues until a dideoxy nucleotide is finally incorporated. This means that the 3'-OH group to form the phosphodiester bond to the next chain link is missing and the synthesis stops here. By defining the starting point, the length of the synthesis fragments in a reaction mixture reflects the relative position of the respective nucleic base type in the molecule. If you use radioactively (or otherwise labeled) nucleotides for the reaction and separate the four approaches next to one another according to their size in an acrylamide gel, you can read the base sequence directly from the gel. The figure opposite illustrates the principle of dideoxy sequencing.
In 1977, Sanger and co-workers presented the complete sequence of the 5,386 base pair bacteriophage φX174. The amino acid sequence of the ten viral proteins could be read directly from the sequence of this bacterial virus, since the genetic code , which specifies which sequence of three nucleobases ( base triplet ) codes for which amino acid in a protein, was known through pioneering work in the 1960s. In 1980 Sanger was awarded the Nobel Prize for the second time for his contributions to the sequencing of nucleic acids.
The importance of Sanger's work for genetic engineering and the genome project
At the beginning of the 1970s, cloning methods were developed with which DNA pieces of any origin can be reproduced in bacteria so that sufficient material is available for sequencing. This opened up the possibility of sequencing the entire genetic information of an organism, the genome , and thus indirectly deriving the sequences of all proteins that can theoretically be synthesized by this organism. Together with Stanley Norman Cohen and Paul Berg , the inventors of the recombinant cloning technique, Frederick Sanger can therefore be described as the father of genetic engineering and the genome project.
Quotes
“Of the three main activities involved in scientific research, thinking, talking, and doing, I much prefer the last and probably best at it. I am all right at the thinking, but not much good at the talking. "
"Previously I had not had much interest in nucleic acids. I used to go to Gordon Conferences on Protein and Nucleic Acids when the two subjects were bracketed together, and would sit through the nucleic acid talks waiting to get back to proteins. However, with people like Francis Crick around, it was difficult to ignore nucleic acids or to fail to realize the importance of sequencing them. "
“Unlike many scientists, I decided to retire and give up research when I reached the age of 65. This surprised my colleagues, and to some extent myself also. I had not thought about retirement until I suddenly realized that in a few years I would be 65 and would be entitled to stop work and do some of the things I had always wanted to do and had never had time for. The possibility seemed surprisingly attractive, especially as our work had reached a climax with the DNA sequencing method and I rather felt that to continue would be something of an anticlimax. "
Works
Important original works:
- For protein sequencing
- F. Sanger: The Free Amino Groups of Insulin. In: Biochemical Journal . Volume 39, No. 5, 1945, pp. 507-515; PMID 16747948 ; PMC 1258275 (free full text).
- F. Sanger: The terminal peptides of insulin. In: Biochemical Journal. Volume 45, 1949, pp. 563-574; PMID 15396627 ; PMC 1275055 (free full text).
- AP Ryle, F. Sanger, R. Kitai: The disulphide bonds of insulin. In: Biochemical Journal. Volume 60, 1955, pp. 541-556; PMID 13249947 ; PMC 1216151 (free full text).
- For DNA sequencing
- F. Sanger, S. Nicklen, AR Coulson: DNA sequencing with chain-terminating inhibitors. In: Proceedings of the National Academy of Sciences . Volume 74, 1977, pp. 5463-5467; PMID 271968 ; PMC 431765 (free full text).
- F. Sanger, GM Air, BG Barrell, NL Brown, AR Coulson, CA Fiddes, CA Hutchison, PM Slocombe, M. Smith: Nucleotide sequence of bacteriophage phi X174 DNA. In: Nature . Volume 265, 1977, pp. 687-695; PMID 870828 .
- Autobiographical, review article
- F. Sanger: Sequences, sequences, and sequences. In: Annual Review of Biochemistry . Volume 57, 1988, pp. 1-28; PMID 2460023 .
- F. Sanger: The early days of DNA sequences. In: Nature Medicine . Volume 7, 2001, pp. 267-268.
literature
- John Walker: Frederick Sanger (1918-2013). In: Nature . Volume 505, 2014, p. 27, doi: 10.1038 / 505027a
- Sydney Brenner: Frederick Sanger (1918-2013). In: Science . Volume 343, No. 6168, 2013, p. 262, doi: 10.1126 / science.1249912
Web links
- Information from the Nobel Foundation on the 1958 award ceremony for Frederick Sanger (English)
- Information from the Nobel Foundation on the awarding of the 1980 award to Frederick Sanger
- Wellcome Trust Sanger Institute
- Autobiography
Individual evidence
- ↑ a b Alok Jha: DNA pioneer Frederick Sanger dies aged 95 ( English ) In: The Guardian . November 20, 2013. Archived from the original on November 21, 2013. Retrieved on November 21, 2013.
- ^ Bavarian Academy of Sciences: Corresponding members . In: badw.de . Bavarian Academy of Sciences. Archived from the original on November 7, 2011. Retrieved November 21, 2013.
- ^ Frederick Sanger: Sequences, Sequences, and Sequences . In: Frederick Sanger, Margaret Dowding (Eds.): Selected Papers of Frederick Sanger: With Commentaries . World Scientific, Singapore / River Edge / London 1996, ISBN 981-02-2430-3 , pp. 635 (Introduction).
- ^ Frederick Sanger: Sequences, Sequences, and Sequences . In: Frederick Sanger, Margaret Dowding (Eds.): Selected Papers of Frederick Sanger: With Commentaries . World Scientific, Singapore / River Edge / London 1996, ISBN 981-02-2430-3 , pp. 648 .
- ^ Frederick Sanger: Sequences, Sequences, and Sequences . In: Frederick Sanger, Margaret Dowding (Eds.): Selected Papers of Frederick Sanger: With Commentaries . World Scientific, Singapore / River Edge / London 1996, ISBN 981-02-2430-3 , pp. 660 .
| personal data | |
|---|---|
| SURNAME | Sanger, Frederick |
| BRIEF DESCRIPTION | British biochemist and Nobel Prize winner in chemistry |
| DATE OF BIRTH | August 13, 1918 |
| PLACE OF BIRTH | Rendcomb , UK |
| DATE OF DEATH | 19th November 2013 |
| Place of death | Cambridge , UK |