chromosome



Bottom: False color representation of all chromosome territories that are visible in this focal plane, according to computer classification.
Chromosomes (from ancient Greek χρώμα chroma , color 'and σώμα Sōma , body') are components of complex (, eukaryotic ') cells on which the inheritance necessary properties of genetic information are stored. One chromosome contains deoxyribonucleic acid (DNA), on which the genes are coded, as well as various proteins , especially histones . In eukaryotic organisms, which include plants , fungi , animals and so also humans , chromosomes are regular components in the cell nucleus .
The term chromosome , literally “color body”, comes from the fact that these structures , which consist of chromatin , can easily be colored with basic dyes.
Without special detection methods, chromosomes can only be recognized by light microscopy during the division of the cell nucleus . Then they have a rod-shaped appearance in humans and many other species. This compressed (condensed) form develops at the beginning of a nucleus division in mitosis like meiosis during the early prophase . In the subsequent metaphase , the strongly condensed chromosomes are arranged equatorially . In this phase, each chromosome consists of two identical chromatids , which were created by replication and each contain a continuous DNA double helix. In the later phases of nuclear division, the two chromatids of a chromosome are separated from each other and finally assigned to the daughter nuclei that are formed.
At the end of the nuclear divisions, the chromosomes return to a decondensed state. Only in this form can the DNA be read or then duplicated . However, the different chromosomes of a nucleus can no longer be differentiated using classic staining methods and form what appears to be a continuous chromatin . With special methods, however, such as fluorescence in situ hybridization , the chromosomes can still be detected as separate units. Each of the decondensed chromosomes thus occupies a definable area within the cell nucleus in the interphase between nuclear divisions, a chromosome territory . In addition to the chromosomes of the cell nucleus, DNA also occurs in some organelles of the cell such as the mitochondria in eukaryotes, and in plants and algae also in the chloroplasts .
The prokaryotic organisms, i.e. bacteria and archaea , do not have a cell nucleus and also have no chromosomes in the classical sense. The carriers of the genetic information here are one or more mostly circular DNA molecules, which are sometimes referred to as bacterial chromosomes . In the mitochondria and chloroplasts of the eukaryotes, the DNA is also usually circular and resembles a bacterial chromosome (see endosymbiont theory ). Your DNA (called chondrioma or plastome ) is sometimes formally listed as an additional, non-nuclear chromosome. The packaging of the long DNA molecules in the smallest space is similar (homologous) to the nucleus of the eukaryotes in archaea , whereas in bacteria it is similar to its organelles (see endosymbiont theory ).
In viruses whose genome consists of one or more nucleic acid molecules (DNA or RNA ), these segments are sometimes referred to as chromosomes. For example, the RNA genome of influenza A viruses consists of 8 such segments (chromosomes).
Research history
The name chromosome was suggested by the anatomist Heinrich Wilhelm Waldeyer in 1888 after Walther Flemming had introduced the term chromatin for the stainable substance in the cell nucleus a few years earlier . In 1906 Oscar Hertwig used the term core segments , which was intended to make it clear that when the core is divided (mitosis) “the chromatin is broken down into segments”. Another old name, which was also used parallel to chromosome for a while , is kernel loop , for example in Karl Heider (1906).
The history of the discovery of chromosomes and their function cannot be separated from the previous discovery of the cell nucleus .
In 1842, the Swiss botanist Carl Wilhelm von Nägeli described “transitory cytoblasts” (stainable rod-shaped structures in the nucleus of plant cells) which were probably chromosomes. With today's knowledge, images from the works of other researchers can also be interpreted as chromosomes or mitotic cell division ( Matthias Schleiden 1846, Rudolf Virchow 1857, Otto Bütschli 1873).

1873 described Anton Schneider of flatworms that the nucleus is feinlockig curved, "in a heap on the addition of acetic acid is transformed visibly expectant threads. Instead of these thin threads, thick strands finally appeared, at first irregular, then arranged in a rosette, which lies in a plane (equatorial plane) going through the center of the sphere. ”The“ indirect nucleus division ”(mitosis) was discovered - but not yet Roger that. In 1882 Walther Flemming assumed that the "core threads" only separate from one another from a previously continuous thread during the early phase of core division. Although he observed a longitudinal splitting of the chromosomes at a later point in time (today referred to as metaphase), he assumed that whole chromosomes (i.e. with both chromatids) later (today: anaphase) move towards the future cell nuclei. Nor did he rule out the possibility that cell nuclei could, at least in some cases, also form anew, i.e. not by dividing from existing nuclei. In 1884 several authors (L. Guignard, Emil Heuser and Edouard van Beneden ) described the division of the chromosome halves (today: chromatids) on the daughter cell nuclei.
Since the chromosomes were not visible during the interphase, it was initially unclear whether they dissolve after a nucleus division and form anew before each nucleus division or whether they survive in the nucleus as separate units. The latter idea was called the doctrine of the preservation of the individuality of the chromosomes and proposed by Carl Rabl (1885). He was also the first to firstly determine a constant number of chromosomes in different mitoses of a tissue and secondly to conclude that the chromosomes must also be present in the interphase and therefore continuously. However, he initially left open the possibility that this number could be different in different tissues. Rabl was also the first to assume that each chromosome in the interphase nucleus forms its own territory.
The idea of chromosome continuity by no means met with unanimous support. Oscar Hertwig (1890, 1917) was an important opponent . Theodor Boveri, on the other hand, endorsed Rabl's ideas and supported them with further experimental findings (1904, 1909). Also in the 1880s, August Weismann developed his germ plasm theory (see also there) , in which he assumed that the genetic material was (only) located in the chromosomes. Important conclusions were that inheritance only takes place via the germline and that inheritance of acquired traits should be rejected. What later proved to be largely correct was hotly controversial at the time. A relentless criticism can be found, for example, in Meyer's Konversations-Lexikon from 1888 under the keyword heredity .
In 1900 the Mendelian rules were rediscovered and confirmed. As a result, the new science of genetics developed , in the context of which the connection between chromosomes and heredity was shown in many ways. For example, in 1910 Thomas Hunt Morgan was able to prove to Drosophila melanogaster that the chromosomes are the carriers of the genes . In 1944 Oswald Avery (see there) showed that the actual genetic molecule is DNA, and not proteins in the chromosomes.
The further story up to 1950 (elucidation of the structure of the DNA) is described in the article Chromosome theory of inheritance . A timeline of some of the important discoveries can be found in the article Chromatin .
In 2000, two international teams of scientists largely deciphered the human genome; in 2003, 99 percent were sequenced. Chromosome 1, the last of the 24 different human chromosomes, was precisely analyzed in 2005/2006 (99.99%). Over 160 scientists from Great Britain and the USA published this joint work.
In 2014 the design and construction of a synthetic chromosome succeeded for the first time, namely in the baker's yeast Saccharomyces cerevisiae .
Structure and structure of the chromosomes
Components

1 One of the two chromatids
2 Centromere , the point where the two chromatids are connected. This is where the microtubules start in mitosis .
3 Short arm (p-arm)
4 Long arm (q-arm)
Apart from special cases (see giant chromosomes below ) a chromosome in the simple case contains a continuous DNA double strand (also: DNA double helix). The double-stranded DNA is also sometimes referred to as the DNA molecule, although, strictly speaking, they are two single-stranded molecules (see deoxyribonucleic acid ) . Histones and other proteins attach to the DNA double strand (see below ). The mixture of DNA, histones, and other proteins is called chromatin . From a DNA double strand annealing protein is through this one chromatid constructed. In this case, the chromosome consists of a chromatid. The case described always occurs immediately after a core division; in most animals and plants also in all cells that can no longer divide (exception: polytene chromosomes in insects, see also below), and in cells that temporarily no longer grow, i.e. are in the G0 phase (see cell cycle ) .
When a cell grows in order to divide later, the DNA has to be doubled (" replicated ") in a certain section of the cell cycle (S-phase) . This is necessary so that both daughter nuclei can later receive the entire genetic material , i.e. one copy of all chromosomes each. After DNA duplication, each chromosome has two identical double strands of DNA. These two double strands are packed spatially separated from each other with proteins: Two sister chromatids are formed. During nuclear division ( mitosis ), the two sister chromatids of a chromosome are microscopically visible as units that run parallel but are separated by a narrow gap (see diagram on the right and first illustration of the article) . At one point, called the centromere or centromere, each chromosome is narrower at this point than in the rest of the process: Here the sister chromatids are still connected. In the further course of mitosis (at the transition from metaphase to anaphase, see below) the two sister chromatids are separated - whereby the separation creates two daughter chromosomes - and distributed to the newly emerging cell nuclei: The chromosomes in these new nuclei now exist again from a chromatid. According to this, a chromatid always contains exactly one DNA double strand, while a chromosome, depending on the phase of the cell cycle, contains one or two DNA double strands and accordingly consists of one or two chromatids (exception: the polytene chromosomes mentioned, which can contain over a thousand double strands).
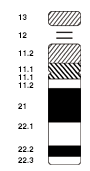
The centromere divides the chromatids into two arms . Depending on the position of the centromere, one speaks of metacentric chromosomes (centromere in the middle), acrocentric chromosomes (centromere at the end, the shorter arm is very small; in humans chromosomes 13 , 14 , 15 , 21 , 22 and the Y chromosome ) or submetacentric chromosomes (centromere between middle and end). The shorter arm is referred to as the p arm ( petit , French for small), the longer the q arm (q follows p in the Latin alphabet). As in the schematic drawing, chromosomes are generally shown with the short arms facing up.
The ends of the chromosomes are called telomeres (singular: telomere). They contain a short, identically repeating DNA sequence (in humans TTAGGG). There the chromosomes become a little shorter each time they are doubled. The telomeres therefore play an important role in the aging process. In addition to centromere and telomeres, starting points for DNA duplication (replication) are the third essential component of a chromosome (see ARS element ) .
In humans, the short arms of the acrocentric chromosomes contain genes for ribosomal RNA . These short arms can be extended in condensed metaphase chromosomes by a satellite , so that satellite chromosomes ( SAT chromosomes ) are present (not to be confused with satellite DNA ). The genes for the ribosomal RNA are present in many copies in tandem, one behind the other. In Interphase - cell nucleus , the forms of these nucleolus . Hence, they are also called nucleolus organizing regions (NOR).
Chromosomes during normal nuclear division (mitosis)
The phases during mitosis are briefly shown below:
- Prophase: In this first stage of mitosis, the chromosomes increasingly condense. They are transformed from an accessible source of genetic information into a compact form of transport that can no longer be read. The nuclear membrane is dissolved. This is sometimes seen as the beginning of an additional phase called the prometa phase .
- Metaphase: The chromosomes migrate to the equatorial plane of the cell and form the metaphase plate there. Up to this point each chromosome consists of two chromatids. Centrioles are located at the opposite poles of the cell; the plasma fibers form the spindle apparatus.
- Anaphase: The spindle apparatus separates the two chromatids of each chromosome and transports them perpendicularly away from the metaphase plate to two opposite cell poles. For this purpose, microtubules are attached to the kinetochores of the centromeres as well as to the cell poles, which act like stretch ropes.
- Telophase: After completion of the anaphase movement, a new nuclear envelope is formed around each individual chromosome and decondensation begins. The fusion of the particles creates the two daughter cell nuclei, which now contain single-chromatid chromosomes.
After the nucleus division, there is usually also cell division, cytokinesis or cytokinesis , which is no longer counted as mitosis.
G, R and other chromosomal bands
In the middle of the 20th century, techniques were developed to "spread" the chromosomes from cells that are in the metaphase: In the resulting metaphase preparation, the chromosomes of a cell lie next to each other on a slide so that they can be counted and linked together under the microscope can be compared (see first figure above) . In well-made preparations, the individual chromosomes have the frequently shown X-like shape. With the classic staining methods such as Giemsa staining , chromosomes are stained evenly over their entire length. For this reason, it was initially difficult or impossible to reliably differentiate between chromosomes of similar size. Around 1970 it was discovered that some areas of the chromosomes no longer accept the Giemsa dye if the chromosomes were previously treated with trypsin . The G banding caused alternating colored sections (the G bands, G for Giemsa) and unstained (the R bands, R for reverse) were created along the chromosomes. The banding pattern enables unambiguous identification of all chromosomes in humans and many animals. The material basis for the different coloring behavior of the bands, i.e. the question of why some areas no longer absorb the dye after trypsin treatment, has not yet been clarified. However, it turned out that G and R bands differ in some properties.

R bands contain an above-average number of genes, an above-average number of GC base pairings and are doubled early during the replication of the chromosomes. In humans they are rich in Alu sequences (see there and figure on the right) .
G-bands are poor in genes, the number of GC base pairs is below average (but they have more AT pairs; see deoxyribonucleic acid ) and they are replicated rather late during the duplication of the chromosomes. In humans they are rich in L1 elements (see Long interspersed nuclear element ) .
C-bands (the centromere regions) and T-bands are sometimes distinguished as further band types. The latter are a subgroup of the R bands, are particularly rich in genes and are often close to the telomeres, hence the name.
The number of R and G bands depends on the degree of condensation of the chromosomes. In the metaphase all human chromosomes together have about 400 of these bands, while in the not yet so strongly condensed prophase chromosomes up to 850 bands can be distinguished.
Nomenclature: In order to enable an exact designation of all chromosomal regions, standardized designation systems have been introduced for humans and some other organisms. In humans, each band has a name made up of the following elements: the number of the chromosome, p or q for the respective arm ( p as in French petite for the short arm; q as in French queue for the long arm ) and numbers, the count up from the centromere. For a finer distinction, the numbers can have several places. The band 3q26.31 is therefore a sub-band of 3q26. The designation “3q” accordingly stands for the entire long arm of chromosome 3. Centromere regions are also designated with c (3c). For the sake of simplicity, telomere areas are often referred to as tel (about 3ptel or 3qtel) and areas close to telomeres with ter (3pter). Schematic representations of the standard bands are called idiograms . Examples are shown in the illustration to the right and on the Ensembl website. In idiograms, G bands are always shown in dark, R bands in white. Areas made up of repetitive elements are sometimes shown hatched. A sorted arrangement of all mitotic chromosomes from a cell is called a karyogram (figure below). The karyotype of a living being indicates how many and possibly which chromosomes this individual has. The karyotype of a woman is given as 46, XX, that of a man as 46, XY (see below, sex determination) .
Size and gene density

The human genome , i.e. the total length of the DNA , comprises around 3.2 Gbp (= gigabase pairs or billions of base pairs) with 23,700 genes found so far. Humans have two copies of the genome (2n), one from the mother and one from the father, which are present in each nucleus. The DNA molecular model results in a length of 3.4 nanometers (billionths of a meter) for 10 base pairs in the double helix . From this it can be extrapolated that the total length of the DNA in every human cell is over 2 meters. In humans, these are distributed over 2n = 46 chromosomes, so that one chromosome contains an average of around 140 Mbp (= megabase pairs or millions of base pairs) and thus a DNA thread of almost 5 cm in length with a little over 1000 genes. However, chromosomes during nuclear division are only a few micrometers (millionths of a meter) in length . They are therefore shortened or “condensed” by a factor of around 10,000. Chromosomes are hardly longer in the interphase nucleus either. The chromosome territories present here are mainly created by the decondensation of the daughter chromatids in width. While a daughter chromatid in the metaphase has a diameter of about 0.6 micrometers, a chromosome territory can occupy a circumference that corresponds approximately to its length. However, chromosome territories can be very irregular in shape. The numerical values given make it clear that chromosomes must be strongly compacted, i.e. unfolded, even during the interphase (see next section) .
Chromosome 1, the largest human chromosome, has 249 Mbp, the shortest chromosome 21 has less than a fifth of it, namely 47 Mbp. The genes are unevenly distributed between the chromosomes. The relatively gene-rich chromosome 19 contains about 1500 coding genes on 59 Mbp, while the gene-poor chromosome 18 on 80 Mbp only contains about 640 (see also figure “Gene-rich and gene-poor regions” above) . However, the poorest is the Y chromosome , which only contains 72 coding genes at 57 Mbp. (Status of the information on sizes and gene densities in this paragraph: December 2015)
In the house mouse ( Mus musculus ) the differences between the chromosomes are smaller. The 3.5 Gbp genome with 22,600 described genes is distributed over 20 different chromosomes (2n = 40) with 61 Mbp (chromosome 19) to 195 Mbp (chromosome 1).
The length of the individual chromosomes in other mammals varies greatly depending on the number. Some have a few, large chromosomes (e.g. the Indian muntjac , Muntjak muntjacus : 2n = 6 in the female and 2n = 7 in the male; so the X chromosome corresponds to two Y chromosomes), others have many small ones (e.g. B. Rhinoceros , Diceros bicornis : 2n = 84). The exact lengths (in base pairs) are only known for a small number of animals.
In lizards and birds chromosomes occur extremely different size on (see figure) . The macrochromosomes are similar in size to mammalian chromosomes. For example, chromosome 1 of the chicken ( Gallus gallus ) contains 188 Mbp. But there are also many microchromosomes whose size can be less than 1 Mbp. The transition from macro to microchromosomes is often fluid, so that the two groups are sometimes differentiated from one another. In the chicken the macrochromosomes can e.g. B. comprise chromosomes 1-8 or 1-10. For a pictorial size comparison see Ensembl. The sizes in Mbp are also taken from there. The terms macro and microchromosomes were introduced by Theophilus S. Painter in 1921, who studied spermatogenesis in lizards.
Molecular structure and hierarchy of the packaging levels
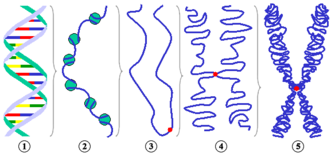
Different levels of chromosome condensation.
1 DNA double helix 2 10 nm fiber ( DNA with nucleosomes ) 3 Schematic chromatin strand during interphase before DNA doubling with centromere 4 Condensed chromatin during prophase (now consisting of two chromatids because the DNA has doubled) 5 Metaphase chromosome Partial figures 3 to 5 are to be understood purely schematically in order to show the number of chromatids during different phases of the cell cycle. The arrangement of the "chromatin thread" does not reflect the actual structure. |

Typical textbook picture of the chromosomal structure with typical errors. See detailed legend here .
|
In the previous section it is shown that the DNA has to be very coiled or “condensed” both during nuclear division and in the interphase . However, it is still largely unclear how this packaging is organized. Basic structural proteins, the histones, play an important role . DNA, histones and other proteins each make up about a third of the chromosomal mass. This is also known as chromatin . The use of the term chromatin is particularly common for descriptions of the cell nucleus in the interphase, as individual chromosomes cannot be distinguished from one another without special staining ( fluorescence in situ hybridization ).
On the lowest packaging level, the DNA thread is wound up in nucleosomes , which contain eight histone molecules (see fig., Sub-picture 2) . Nucleosomes have a diameter of about 10 nanometers (nm), which is why one speaks here of the 10-nm fiber. Their structure is often compared to a pearl necklace, where the thread is wrapped around the pearl. 146 base pairs of DNA are wound up in a nucleosome, plus linker DNA between the nucleosomes. The 10 nm fiber can be detected in the electron microscope, as can the next higher packaging level, the 30 nm fiber. The internal structure of the 30 nm fiber, i.e. how it is assembled from the 10 nm fiber by being unfolded, is just as unclear as all the higher packaging levels. Various models are discussed for the latter. In the loop model (of English. Loop "loop"), it is assumed that the 30-nm fiber runs in large loops, which are fixed to a kind of backbone. In the Chromonema model, on the other hand, it is assumed that the 30 nm fiber is thickened by further unfolding, resulting in sections 120 nm and thicker. How the structural change from the interphase to the prophase chromosome takes place is also unclear. When it comes to the transition from the prophase chromosomes to the even more condensed metaphase chromosomes, there seems to be agreement that this is a spiral-shaped winding.
The condensation of chromosomes or chromatin is not uniform within the cell nucleus. Some areas of the nucleus are particularly strongly colored by DNA dyes. The condensation is particularly strong here. These areas are called heterochromatin , whereas less colored areas are called euchromatin . In the more condensed areas, gene activity is hindered or blocked, see epigenetics .
Giant chromosomes
Two types of giant chromosomes are known, polytene chromosomes and lamp brush chromosomes .
Polytene chromosomes
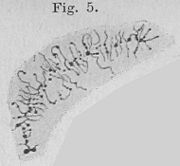
The polytane chromosms represent a special feature with regard to the internal chromosomal structure. They are known from various insects and have been studied particularly well in the fruit fly Drosophila melanogaster and in Chironomus . They arise from several rounds of DNA duplication without subsequent nuclear division (endoreduplication). In contrast to “normal” polyploidy , in polytene chromosomes the multiple replicas of DNA threads from both homologous chromosomes (ie the copy inherited from the father and the copy inherited from the mother) are arranged in parallel, similar to a cable strand. All copies of a gene are therefore next to each other.
Lamp brush chromosomes
Another form of very large chromosomes is found in the ova of amphibians. Since they resemble a bottle or lamp brush from the microscopic point of view, they were called lamp brush chromosomes.
| Polytene chromosomes in a Chironomus salivary gland cell . Walther Flemming , 1882. | "Chromatic thread, which is comparable to a bottle brush" (according to today's terminology a lamp brush chromosome) from the nucleus of an egg cell of the water salamander ( Triton ). ( Entire panel. ) Oscar Hertwig , 1906. |
Sex determination by chromosomes and their consequences
While some living beings are sexed by environmental conditions such as temperature during embryonic development, others are sexed by inherited chromosomes: they have a chromosomal sex . Different groups of animals have produced different methods of determining chromosomal sex, and in some cases similar systems have been developed independently of one another. In mammals and some other groups of animals, females have two X chromosomes , while males have one X and one Y chromosome . If there are two different sex chromosomes, as in male mammals, one speaks of hemizygosity . In birds, males have two Z chromosomes , females are hemizygous with a Z and a W chromosome. Many insects from the group of Hymenoptera females are diploid , the males but only haploid .
In the hemizygous sex, many genes are only present on one chromosome. If there is a genetic defect, it cannot be picked up by an intact gene on a homologous chromosome. Therefore, there are a number of hereditary diseases in humans that practically only occur in men. The most famous examples are a form of hemophilia , Duchenne muscular dystrophy and red-green blindness .
With chromosomal sex determination, one chromosome is present twice in one of the sexes, while the other only has one. In order to prevent that twice as much gene product is produced here as in the opposite sex, different groups of animals have developed different strategies for "dose compensation" (see sex chromosome , X-inactivation and sex chromatin ) .
Chromosome number
Karyotype: The chromosomes of an individual
All the different chromosomes that occur in an individual together make up the karyotype . In many cases (also in mammals ) there are always two homologous chromosomes in the karyotype, apart from the sex chromosomes in the hemizygous sex , which as such carry the same genes. In these cases one speaks of a double or diploid set of chromosomes , which is abbreviated with 2 n . In the case of sexually reproducing organisms, one was inherited from both parents.
The individuals of the same species and of the same sex usually have the same set of chromosomes and thus the same karyotype. An exception are so-called B chromosomes , which occur in some species and can be present in different numbers in different individuals and in different body cells.
Even with the regular chromosomes of a species, there can be differences between the sexes in terms of shape and - less often - number of chromosomes. The sexes then have a different karyotype (see above, sex determination). Humans, for example, have 46 chromosomes in both sexes, but the Y chromosome is smaller than the X chromosome. The karyotype is given as 46, XX for women and 46, XY for men. Karyotypes are determined with the help of karyograms (see below).
Passing on the chromosomes to the next generation
In order to prevent an increase in the number of chromosomes from generation to generation during fertilization, the number of chromosomes in the cell nucleus must be reduced before mature germ cells are formed . As a reduction division, this is a component of meiosis . During meiosis occurs by crossing over to a recombination of homologous chromosomes. This creates genetically newly composed chromosomes that differ from those of the parent organisms. During the division, it is a matter of chance which of the recombined chromosomes together form a single set of chromosomes in the nucleus of the resulting haploid cells. The chromosome segments that were previously inherited on the paternal and maternal side come together in different combinations in the new haploid chromosome set of germ cells.
In diploid animals , haploid germ cells are produced, egg cells or sperm . The germ cells can fuse and become the first cell of a new living being, the zygote . The two simple chromosome sets in the two pronuclei then become the double chromosome set in the cell nucleus . In hybridogenesis , a deviation from a random distribution of chromosomes, which has been found in a few animal species, occurs.
In plants and protozoa , haploid and diploid generations can alternate (see generation change ). Sometimes the diploid status is very short-lived and the haploid generation predominates.
Non-diploid sets of chromosomes
Occasionally there is the opinion that all higher animals and plants have a double set of chromosomes, i.e. are diploid. However, this is not the case. Although the majority of animals and many plants are diploid, there are quite a few with different degrees of ploidy .
As mentioned above, haploid individuals occur when the generation of plants changes. In addition, haploid males are found in a number of insect species ( haplodiploidy ) and probably also in some mites . One case of haploid female animals is also known: the mite species Brevipalpus phoenicis , a pest of tropical crops, consists only of haploid females that reproduce parthenogenetically . According to one study, it is actually genetic males that become females when infected with bacteria . Feminization through bacterial infection is also known in other arthropods , mostly through Wolbachia .
In some species there are sets of chromosomes that are composed of more than two simple ones. These, too, are not two-fold, diploid, but show higher degrees of ploidy. With 3 n they are referred to as triploid , with 4 n as tetraploid , with 6 n as hexaploid etc., or generally as polyploid . In plants, usually the number is on chromosomes in the haploid genome of an organism radix or base number called and x respectively. If the number of chromosomes is an integral multiple of the base number, it is called euploidy . Diploid plants then have 2 x chromosomes, tetraploid 4 x etc. The genome of a tetraploid plant, for example with the base number x = 7, then has 4 x = 28 chromosomes.
Tetraploidy is probably the second most common degree of ploidy after diploidy. It has been observed in many flowering plants , insects and also amphibians . Tetraploidy can come by after DNA replication and Chromatidenverdopplung a cell division is prevented. Many crops, e.g. B. in the cereals, created by polyploidization from diploid wild forms.
Even higher degrees of ploidy occur in plants. For example, they can arise when two species are crossed and the children keep all the chromosomes from their parents. One then speaks of addition bastards . Modern seed wheat , for example, is hexaploid .
Triploid individuals can arise when diploid and tetraploid individuals mate. This is possible when both are closely related species. However, triploid individuals will usually be sterile, as the odd number of more simply composed chromosomes result in difficulties in pairing the chromosomes during meiosis . Exceptions, that is, triploid individuals capable of reproduction, were discovered in amphibians . Sometimes diploidy, tetraploidy and also triploidy occur in closely related species or in the same species next to each other. In the water frog , one of the inherited simple sets of chromosomes is specifically eliminated before entering meiosis (see also pond frog ). A localized, triploid population of green toad has been found in Pakistan and this can also be observed.
In theory at least, there can be a smooth transition from tetraploid to diploid, for example. In a tetraploid living being, all chromosome pairs are duplicated. Changes in one of the two pairs, for example the loss of individual genes, can therefore be tolerated. The gene copies on the two pairs can also develop apart during further evolution and take on different functions. Chromosome mutations ( see below ) in only one of the two pairs are also possible. If many such changes come together in the course of time, the originally identical pairs of chromosomes have developed so far apart that one can no longer speak of four-fold chromosome sets: Now diploidy is again present. For the early history of the vertebrate genesis , two rounds of such genome duplications have been proposed (“2R hypothesis”), with which today's diploid vertebrates would have evolved from originally octaploid (8 n ) organisms. This would explain why, for example, the Hox gene clusters occur four times per haploid genome of vertebrates, but only once in other animals.
The degree of ploidy of individual body cells of a multicellular cell can deviate from the degree of ploidy of the organism. The best-known example of this are the polytene chromosomes of some insects (see above). However, in addition to the predominant diploid cells, haploid, triploid and tetraploid cells have also been described in rare cases for the rat liver. Tetraploidy is easily caused by duplication ( reduplication ) of the chromosomes without subsequent nuclear division, i.e. by endoreduplication or endomitosis . Haploidy and triploidy in body cells of diploid organisms have been reported so rarely that experimental errors or artifacts cannot be ruled out. The potential mechanism of formation is unclear. High degrees of ploidy are associated with correspondingly larger cell nuclei. Due to the larger amount of genetic material, very large body cells can also be supplied.
Table: Number of chromosomes in normal body cells
Unless otherwise stated, the figures are based on Flindt, 1985.
Mammals
|
Other vertebrates
|
Invertebrates
|
Other eukaryotes
|
Karyogram
A karyogram is a sorted representation of the chromosomes of a metaphase preparation. These preparations are made by spiking cell cultures with an agent that prevents the formation of microtubules , e.g. B. colchicine or nocodazole . As a result, no spindle apparatus can develop and the cell cannot go into anaphase. As a result, a number of cells accumulate in the metaphase (see above ) and the yield is increased accordingly. The cells are treated hypotonically , causing them to swell, fixate and drop onto a slide, causing the metaphase chromosomes to lie next to each other (see first picture above) . The chromosomes are stained, photographed and arranged according to size in the karyogram so that the karyotype can be determined (see figure on the right) .
Karyograms are used both in the investigation of the karyotypes of organisms and in clinical application when chromosomal changes are suspected.
Chromosome mutations
Permanent changes to the chromosomes can occur when breaks occur in at least two places in the DNA double helix. In most cases, DNA double-strand breaks are properly repaired so that permanent changes do not occur. However, if the wrong ends of two different breaks are joined during a DNA repair , chromosome mutations occur. If the breakpoints are on the same chromosome, deletions (loss of a section) or inversions (flipping) can occur. Another type of mutation within a chromosome is duplication (duplication of a section). If the double-strand breaks are on different chromosomes, translocations can occur. These phenomena are described in more detail in their own articles.
Chromosome mutations play a role both in chromosome evolution and in the clinical field. In terms of clinical significance, hereditary diseases (see also below) , tumor development (e.g. the Philadelphia chromosome ) and radiation biology should be mentioned.
A distinction must be made between numerical changes, i.e. an additional or a missing chromosome, from the structural changes mentioned. These are not referred to as a chromosome mutation. Since only a single chromosome is affected, one speaks of trisomy ( not triploidy) or monosomy (see chromosome aberration ) .
Chromosome evolution
The change in chromosomes in the course of evolution is called chromosome evolution . Similar to external physical characteristics or the sequence of individual genes , the tribal history can also be traced on the chromosomes . For example, the chromosomes of humans (46 pieces) are very similar to those of the great apes ( chimpanzees , gorillas and orangutans , each with 48 chromosomes). There are only two inter-chromosomal tags within this species group. Chromosome 2 is specifically human. The other species mentioned have two smaller chromosomes instead of this, which contain the corresponding gene sequences (see illustration) . Gorilla-specific, on the other hand, is a translocation between those chromosomes that correspond to human chromosomes 5 and 17. This results in the original karyotype of the group with 48 chromosomes, as it is still present in chimpanzees and orangutans today.

An evolutionarily stable change in the chromosomes is only possible if a chromosome mutation occurs in the germline . A “balanced” change, in which all chromosome segments are present in the correct number, initially has no disease value for the carrier. However, there are difficulties in meiosis . The change initially only occurs on one chromosome (or on two in the case of fusions or translocations), but not on the respective homologous chromosomes. Since unlike otherwise identically structured partners, there is no normal meiotic mating. The risk of segregation errors and the resulting germ cells with redundant or missing chromosomal segments (and consequently sick children) increases sharply. In the vast majority of cases, such changes will therefore be lost again in the following generations. A stable situation is only achieved if both copies of the chromosomes involved carry the corresponding change. This could happen, for example, if a dominant male with a change has numerous children, who in turn mate with one another, so that grandchildren with the change arise on both copies of the chromosomes involved. These offspring do not have a selection disadvantage when they mate with one another. When mating with individuals with the original chromosomes, however, the resulting children again experience reduced fertility due to segregation errors. It is therefore assumed that “fixed” chromosome changes are a mechanism for speciation .
More closely related species or groups of species do not always have to have more similar chromosomes than more distant species. For example, the chromosomes of great apes, including humans, are very similar to those of macaques ( Macaca fuscata ). However, the chromosomes of the closely related small great apes ( gibbons ) differ greatly from both those of the great apes and those of the macaques. Due to numerous modifications, only five of the gibbon chromosomes are (only) homologous to one human chromosome over their entire length. Obviously, evolutionary changes in the karyotype proceed much faster in some groups (e.g. the gibbons) than in others (macaques, great apes). It is assumed that this is not due to a higher mutation rate, but to a more frequent fixation of changes that have occurred. One reason for this could be different lifestyles or social behavior. Gibbons live in small groups in which chromosome changes could prevail faster than in large herds. In Gibbons there are chromosomal polymorphisms (differences) in the karyotype of examined animals of the same species, which indicate that the rapid chromosome evolution continues in this group of animals. However, the relatively large number of polymorphisms also suggests that the selective disadvantage of mixed forms may be less than originally thought.
Chromosomes in humans
Humans have 46 chromosomes, 2 of which are sex chromosomes or gonosomes (XX in women, XY in men, see above: sex determination ). The chromosomes of the remaining 22 chromosome pairs are called autosomes . The autosomes were numbered from 1 to 22 according to their size in the microscopic preparation. Like other mammals , humans are diploid , so a cell has a double set of chromosomes : there are two copies of chromosomes 1 to 22, plus the two sex chromosomes.
Properties of the sex chromosomes
Although the X chromosome with 155 megabases and the Y chromosome with 59 megabases differ greatly in size, they also have something in common. At both ends they contain regions in which the DNA sequence between the X and Y chromosomes is very similar, the pseudoautosomal regions (PAR). There are several genes in the PARs, which are duplicated in both sexes and which are not subject to X inactivation (see above: dose compensation ) . In these regions during which meiosis is a recombination between X and Y chromosome possible.
Even in non-recombining regions of the Y chromosome, around half of the genes have equivalents on the X chromosome. These are mainly genes of the basic metabolism . Two of the genes that are also found on the X chromosome are only active in the testicle . The other genes without a counterpart on the X chromosome are also only active in the testes, determine the male gender and control sperm production. Loss of a piece of the long arm near the centromere leads to short stature.
Genome and chromosome mutations of clinical significance
By chromosome aberrations , so chromosomal mutations , chromosomal instability , chromosomal breakage or an incorrect number of chromosomes (numerical chromosome aberrations or genome mutation ), it can lead to clinical syndromes with severe partly symptoms occur.
It is not always possible to assign the clinical pictures to either chromosome mutations or numerical chromosome aberrations. So z. B. Down's syndrome is caused in most cases by an additional, complete chromosome 21 (free trisomy ). However, around 3% of cases are due to translocations in which part of chromosome 21 is fused to another chromosome. Only this part is then present three times. The following syndromes are mostly dealt with in detail in their own articles and are only presented here as an overview.
Autosomal trisomies
Free trisomies in live births are only known for chromosomes 21, 18 and 13 in the autosomes. All three belong to the gene-poor chromosomes (compare the second figure in the section G, R and other chromosome bands above). From this it can be concluded that free trisomies of the other autosomes are incompatible with life.
- Down syndrome or trisomy 21 (triple / trisomal presence of genetic material of chromosome 21 in all or some body cells). Occurrence: 1 case in 600–800 newborns. Important symptoms include a. Heart defects and mental retardation. Whereas in the past most of those affected died of infectious diseases in childhood , the average life expectancy is now over 60 years.
- Edwards syndrome or trisomy 18 (triple / trisomal presence of genetic material of chromosome 18 in all or some cells of the body). Occurrence: 1 case in 2,500 newborns. Organ malformations are diverse, u. a. Heart defects and kidney malformations. Severe intellectual disability (no language), adulthood is only reached in exceptional cases.
- Patau syndrome or trisomy 13 (triple / trisomal presence of genetic material of chromosome 13 in all or some body cells). Occurrence: 1 case in 6,000 newborns. Common symptoms include a. Heart defects, cleft lip and palate , polydactyly (multiple fingers) and severe intelligence defects . Adulthood is only reached in exceptional cases.
- Trisomy 8 (threefold / trisomal presence of genetic material of chromosome 8 in some body cells). Common symptoms include a. deep hand and foot lines, vertebral malformations, neural tube malformations (often spina bifida aperta ) and tall stature.
Variations in the number of sex chromosomes
- Ullrich-Turner syndrome (45, X). Missing second sex chromosome. Occurrence: 1 case in 3,000 newborns. Women with this syndrome have underdeveloped female sexual characteristics, short stature, deep hairline, unusual eye and bone development, a funnel-shaped chest, and are mostly sterile. The intelligence is normal, sometimes spatial imagination or math skills are below average.
- Triplo-X syndrome (47, XXX). Triplo-X syndrome is the most clinically normal chromosomal aberration. Presumably many cases are never discovered. Intelligence is usually lower than that of siblings. Fertility can be slightly reduced. The offspring show a barely increased rate of chromosome aberrations.
- 48, XXXX and 49, XXXXX. As the number of X chromosomes increases, intelligence and fertility decrease.
- Klinefelter syndrome (almost always 47, XXY; rarely 48, XXXY or 49, XXXXY). 1 case in 1,000 male newborns. Men with this syndrome are often sterile, tall, have unusually long arms and legs, a tendency to develop breasts (pseudo- gynecomastia ), and reduced body hair. The intelligence quotient is on average 10 lower than that of siblings.
- XYY syndrome (47, XYY). Men with this syndrome are usually phenotypically normal and diagnosed by chance. Life expectancy is not restricted, fertility is almost normal, they are on average 10 cm taller than their brothers and their intelligence is slightly reduced compared to siblings. Occasionally, disorders associated with chromosome aberration such as undescended testicles can occur.
- Higher grade Y polysomies: 48, XXYY men are similar to XYY men, but sterile and with a tendency to be less intelligent. The latter increases in 48, XYYY and the very rare 49, XYYYY men. Organ malformations also occur.
Marker chromosomes
Marker chromosomes are all chromosomes that are not readily definable and that appear in addition to normal chromosomes. They consist of material from the normal chromosomes, but are usually small so that they cannot be identified by G-banding (see above ) . This can be achieved with high resolution fluorescence in situ hybridization .
Deletions on autosomes
Autosome monosomies do not occur. The associated damage is apparently incompatible with life. However, there are a large number of different deletions of parts of an autosome, some of which are only known from a few clinical cases. The following list is therefore not exhaustive and only includes the most popular examples.
- Although not long known, deletion of the end of the short arm of chromosome 1 is probably the most common deletion (1 case in 5,000-10,000 newborns). The symptoms are not uniform, mostly severe mental retardation is present.
- The Cri-du-chat syndrome (cri du chat) is deletion caused the end of the short arm of chromosome 5th It was described as the first autosomal deletion in 1963. The frequency is around one case in 50,000 newborns. In early childhood, children stand out for their high-pitched screaming, which is reminiscent of the screaming of cats and which is caused by malformations of the larynx . They have eyes that are wide apart ( hypertelorism ), a small head ( microcephaly ) and jaws, and their intelligence is reduced . Since internal organs are usually not affected, the chances of survival are comparatively good.
- The Wolf-Hirschhorn syndrome is deletion caused the end of the short arm of chromosome. 4 The frequency is also around one case in 50,000 newborns. Affected people are usually severely impaired cognitively and have stunted growth. Less than half of the children survive the first 18 months.
- The De-Grouchy syndrome occurs in two variants of chromosome 18 caused by deletions of various arms.
Further examples are the Williams-Beuren syndrome (7q11.23) and the Smith-Magenis syndrome (17p11.2 - frequency between 1: 15,000 to 1: 25,000 births stated).
Deletions of the region 15q11.2-q12 represent a specialty. This region is subject to an epigenetic regulation, the " imprinting ": Depending on whether this region was inherited from the father or the mother, certain genes are active or inactive. Usually both cases exist once. If one of the two is missing, e.g. B. by deletion, the clinical pictures differ depending on whether a region inherited from the mother ( Angelman syndrome ) or a region inherited from the father ( Prader-Willi syndrome ) is missing.
The ICD-10 code O35.1 is given when caring for an expectant mother in the event of (suspected) chromosomal peculiarities in the unborn child.
literature
- Gholamali Tariverdian, Werner Buselmaier: Chromosomes, genes, mutations. Human Genetic Consultation . Springer, Berlin 1995, ISBN 3-540-58667-9 .
- Walther Traut: Chromosomes. Classical and Molecular Cytogenetics . Springer, Berlin 1991, ISBN 3-540-53319-2 .
- Jan Murken, Tiemo Grimm, Elke Holinski-Feder: Pocket textbook human genetics . 7th edition. Thieme, Stuttgart 2006, ISBN 3-13-139297-5 .
- Thomas Cremer: From cell theory to chromosome theory. Springer Verlag, Berlin / Heidelberg / New York / Tokyo 1985, ISBN 3-540-13987-7 . (on-line)
- Bärbel Häcker: Chromosomes. In: Werner E. Gerabek , Bernhard D. Haage, Gundolf Keil , Wolfgang Wegner (eds.): Enzyklopädie Medizingeschichte. De Gruyter, Berlin / New York 2005, ISBN 3-11-015714-4 , p. 261 f.
Web links
- Lexicon of biology : chromosomes
- Chromosomes; Chromosome Theory (Part II)
- Chromosome structures and structural changes in the chromosomes
- Fine structure of the chromosomes
- Thumbnail overview chromosomes
- Human chromosome morphology
- Chromosome numbers for the flora of Germany
Individual evidence
- ^ Wilhelm Gemoll : Greek-German school and hand dictionary. Munich / Vienna 1965.
- ↑ Bärbel Häcker: Chromosomes. 2005, p. 261.
- ↑ retrobibliothek.de
- ↑ Gregory et al: The DNA sequence and biological annotation of human chromosome. In: Nature . 441, 1, 2006, pp. 315-321. doi: 10.1038 / nature04727
- ↑ N. Annaluru et al.: Total synthesis of a functional designer eukaryotic chromosome. In: Science . tape 344 , no. 6179 , April 4, 2014, p. 55-58 , doi : 10.1126 / science.1249252 .
- ↑ D. Schindler, T. Waldminghaus: Synthetic chromosomes. In: FEMS Microbiol Rev . 2015, doi : 10.1093 / femsre / fuv030 , PMID 26111960 .
- ^ Graw, Jochen .: Genetics . 6th, revised and updated edition. Springer Berlin Heidelberg, Berlin, Heidelberg 2015, ISBN 978-3-662-44817-5 .
- ↑ a b c Homo sapiens . on ensembl.org (English) database from September 2006.
- ↑ Mus musculus (house mouse) on ensembl.org (English). Database status from March 2017.
- ↑ a b Gallus gallus (chicken). on ensembl.org (English) database from September 2006.
- ^ Theophilus S. Painter: Studies in reptilian spermatogenesis . I. The spermatogenesis of lizards. In: Jour Exper Zoology. 34, 1921, pp. 281-327.
- ↑ AS Belmont, K. Bruce: Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. In: J. Cell Biol. 127, 1994, pp. 287-302.
- ↑ Panagiota Manolakou, Giagkos Lavranos, Roxani Angelopoulou: Molecular patterns of sex determination in the animal kingdom: a comparative study of the biology of reproduction. In: Reproductive Biology and Endocrinology. 4, 2006, p. 59. doi: 10.1186 / 1477-7827-4-59
- ↑ Andrew R. Weeks, Frantisek Marec, Johannes AJ Breeuwer: A mite species that consists entirely of haploid females. In: Science . 292, 2001, pp. 2479-1482. doi: 10.1126 / science.1060411
- ↑ Basic number in the lexicon of biology on Spektrum.de
- ^ Peter Sitte , Elmar Weiler , Joachim W. Kadereit , Andreas Bresinsky , Christian Körner : Textbook of botany for universities . Founded by Eduard Strasburger . 35th edition. Spektrum Akademischer Verlag, Heidelberg 2002, ISBN 3-8274-1010-X , p. 531 .
- ↑ Matthias Stöck, Dunja K. Lamatsch, Claus Steinlein, Jörg T. Epplen, Wolf-Rüdiger Grosse, Robert Hock, Thomas Klapperstück, Kathrin P. Lampert, Ulrich Scheer, Michael Schmid, Manfred Schartl: A bisexually reproducing all-triploid vertebrate. In: Nat. Genet. 30, 2002, pp. 325-328. doi: 10.1038 / ng839 .
- ↑ Paramvir Dehal, Jeffrey L. Boore: Two rounds of whole genome duplication in the Ancestral Vertebrate. In: PLoS Biol . 3 (10), 2005, p. E314. doi: 10.1371 / journal.pbio.0030314
- ↑ Eberhard Gläss: The identification of the chromosomes in the karyotype of the rat liver. In: Chromosoma. 7, 1955, pp. 655-669. doi: 10.1007 / BF00329746
- ↑ a b c Rainer Flindt: Biology in numbers . A data collection in tables with over 9000 individual values. Gustav Fischer, Stuttgart 1985, ISBN 3-437-30466-6 , pp. 86 f., 138 f .
- ↑ a b c d e f g h chimpanzee on ensembl.org (English) database status from September 2006.
- ↑ a b c d e A. Jauch, J. Wienberg, R. Stanyon, N. Arnold, S. Tofanelli, T. Ishida, T. Cremer: Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. In: Proc Natl Acad Sci USA. 89, 1992, pp. 8611-8615. PMC 49970 (free full text)
- ↑ a b Jaclyn M. Watson, Julianne Meynedagger, Jennifer A. Marshall Graves: Ordered tandem arrangement of chromosomes in the sperm heads of monotreme mammals. In: Proc. Natl. Acad. Sci. UNITED STATES. 93, 1996, p. 10200.
- ↑ a b Theodor Boveri : The potencies of the Ascaris blastomeres with modified furrowing. Publishing house by Gustav Fischer, Jena 1910.
- ↑ GJ Hunt, RE Page Jr: Linkage Map of the Honey Bee, Apis Mellifera, Based on Rapd Markers. In: Genetics. 139 (3), 1995, pp. 1371-1382, Table 1. PMC 1206463 (free full text).
- ^ R. Vitturi, A. Libertini, L. Sineo et al .: Cytogenetics of the land snails Cantareus aspersus and C. mazzullii (Mollusca: Gastropoda: Pulmonata) . In: Micron . tape 36 , no. 4 , 2005, p. 351-357 , doi : 10.1016 / j.micron.2004.12.010 , PMID 15857774 .
- ↑ V. Wendrowsky: About the chromosome complexes of Hirudinea. In: Journal of Cell Research and Microscopic Anatomy. (now: Cell and Tissue Research) 8 (2), 1928, pp. 153-175. doi: 10.1007 / BF02658704
- ↑ Ensembl database , accessed on January 24, 2014.