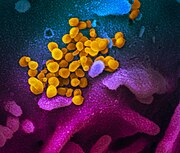
SARS-CoV-2
The virus SARS-CoV-2 (abbreviation for English severe acute respiratory syndrome coronavirus 2 , German severe acute respiratory syndrome coronavirus-2 ), colloquially just called (new) coronavirus , belongs to the coronavirus family . An infection with this virus, the new respiratory disease COVID-19 cause.
The SARS-CoV-2 virus was first discovered in the city of Wuhan ( People's Republic of China ) at the end of 2019 and triggered the COVID-19 pandemic worldwide . The World Health Organization (WHO) classified it on January 30, 2020 as a “ health emergency of international concern ” and on March 11, 2020 as a pandemic .
The infection usually occurs via droplet infection in close contacts and via aerosols also in social interaction. Transmission through smear infection cannot be ruled out. The spreading takes place in particular through so-called superspreading .
Discovery story
In December 2019, an accumulation of severe pneumonia with an unknown cause was found in Wuhan City . On December 30, 2019, the Chinese doctor Li Wenliang in a WeChat group informed his doctor colleagues about seven patients who were treated with suspected infection with the SARS virus in the Wuhan Central Hospital; he was warned for this by the Chinese police. Li Wenliang later developed COVID-19 himself and died as a result. The Chinese Center for Disease Control and Prevention sent a team to the city on December 31, 2019. On the same day, the China office of the World Health Organization (WHO) was officially informed by the Chinese authorities that several people in Wuhan had developed severe pneumonia in December 2019 and that the cause of this was suspected to be a previously uncharacterized, infectious pathogen . As of January 3, 2020, a total of 44 sick people were reported to the WHO, including several seriously ill. Since several sick people worked as sellers or traders at the local wet market “Wuhan Huanan Wholesale Market for Fish and Seafood” ( Chinese 武汉 华南 海鲜 批发市场 , Pinyin Wǔhàn huánán hǎixiān pīfā shìchǎng ), this market was suspected to be the primary infection site. Shortly after the onset of the disease in December 2019, 27 or 66% of the first 41 hospital patients in Wuhan had visited the market in the city center. However, according to this study, the infections of 13 of the other affected people were in no way related to this location. On January 7, 2020, the head Chinese virologist Xu Jianguo ( 徐建国 ), who is involved in virus identification, announced that the pathogen was a previously unknown coronavirus. This would have shown examinations of blood samples and throat swabs from 15 sick people. This finding was confirmed in a WHO statement on January 9, 2020. On January 13, 2020, the complete genome sequence of an isolate of the new coronavirus was deposited in the NCBI - GenBank (GenBank number MN908947). A first verification procedure was published almost at the same time.
A phylogenetic analysis of the genome sequences of environmental samples (e.g. from surfaces) on the market that has now been carried out showed that they are very closely related to the viruses of the first patients from Wuhan. According to a study by the Wuhan Hospital, however, the first patient identified had not visited the market. None of the examined animals from the market tested positive for SARS-CoV-2, which supports the assumption that the virus did not spread to humans there. Apparently, the virus had previously established itself among people unnoticed. Instead, the market could have been the site of an early superspreader event; similar to how it later appeared in various other places around the world. A 55-year-old man from Hubei province is now suspected of being patient zero, who could have been infected on November 17, 2019. In the meantime, a case in France near Paris was detected in December 2019 by retrospective analysis. The patient had no connection with China / Wuhan, but his wife worked in a supermarket near the airport. There is also an even earlier suspicion of patient zero in Alsace / France on November 16, 2019.
So it turned out that not all early COVID-19 cases can be linked to the market and the history of the outbreak is arguably more complicated than originally assumed.
designation
In public, the "SARS-CoV-2" virus is usually (after the virus family) as "novel coronavirus", "new coronavirus", "coronavirus", just "corona" or occasionally (after the illness) as "COVID-19 -Virus ”.
The designation "2019-nCoV" used by the World Health Organization (WHO) from January 13 to February 11, 2020 was only considered to be "provisional" according to the WHO. The first sequenced virus isolate was designated in the first description as WH-Human-1 coronavirus (WHCV) (WH = Wuhan) and as Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1 in the NCBI taxonomy database (the one for virus names and - classifications is not decisive). The virus was then listed there - also temporarily - as the Wuhan seafood market pneumonia virus ; as synonyms were 2019 Ncov and Wuhan coronavirus mentioned. The WHO did not take up various proposed names, all of which had in common, the virus - based on MERS-CoV ( Middle East respiratory syndrome coronavirus ) - after the place of its initial identification (the city of Wuhan) as Wuhan respiratory syndrome coronavirus (WRS-CoV) to name. One reason given by experts was complaints in the past, when viruses were named after individual countries or regions (for example the Marburg virus ). The WHO therefore issued recommendations in 2015 on how to name new pathogens and diseases. Naming after the place of discovery was explicitly declared undesirable. Synonyms listed in the NCBI taxonomy database are 2019-nCoV , COVID-19 , COVID-19 virus , Wuhan coronavirus and Wuhan seafood market pneumonia virus (as of February 16, 2020).
The coronavirus SARS-CoV-2 was also named "2019-nCoV" ("2019 novel coronavirus"), "novel coronavirus 2019" and "Wuhan coronavirus" prior to being named by the International Committee on Taxonomy of Viruses (ICTV).
On February 11, 2020, the WHO announced that it had named the disease caused by the virus as "COVID-19" or "Covid-19" (for corona virus disease 2019 ). On the same day, the Coronavirus Study Group (CSG) of the International Committee on Taxonomy of Viruses (ICTV) proposed the name SARS-CoV-2 (for severe acute respiratory syndrome coronavirus 2 ) for the virus on the preprint server bioRxiv . A week later, a group of Chinese virologists contradicted this and instead suggested HCoV-19 ("Human Coronavirus 2019") as the name human coronavirus 2019 . As a reason, they cited the similarity of this virus name with the name of the disease named by the WHO as COVID-19 and the risk of confusing SARS-CoV-2 with SARS-CoV in public. They emphasized that "2019-nCoV" is different from the SARS virus in biological and epidemiological terms, as are the clinical symptoms of COVID-19 and SARS. To differentiate, the SARS pathogen is also referred to as SARS-CoV-1.
features
Systematics
SARS-CoV-2 is one of the representatives of the species Severe acute respiratory syndrome-related coronavirus (SARS-associated coronavirus, acronym SARSr-CoV). Currently (March 2020) only SARS-CoV-2 and SARS-CoV-1 belong to this species. The latter was previously known simply as SARS-CoV and is the causative agent of SARS , while SARS-CoV-2 triggers COVID-19 .
The species Severe acute respiratory syndrome-related coronavirus is currently the only species in the subgenus Sarbecovirus . The name "Sarbecovirus" refers to the English name SAR S-like be ta co ronavirus (or SAR S-related be ta co ronavirus ).
The subgenus Sarbecovirus belongs to the current genus Betacoronavirus . The previous genus Coronavirus has been abolished and its members have been divided into the new genera Alpha- , Beta, Gamma and Delta Coronavirus . Today, the name "Coronavirus" can informally designate all viruses of the Coronaviridae family and is used especially for the general public for easier understanding.
The MERS-CoV virus, which is often referred to in connection with SARS-CoV-2 , also belongs to the Betacoronavirus genus , but to the Merbecovirus sub-genus there .
The genus Betacoronavirus belongs to the subfamily of the Orthocoronavirinae (previously simply Coronavirinae ), and this to the family of Coronaviridae, this to the suborder Cornidovirineae , this to the order Nidovirales . The latter is classified in the virological area of RNA viruses or riboviruses ( riboviria ) , since their genetic material consists of RNA . However, this does not express any further relationships in the phylogenetic sense.
A family tree of the SARS-CoV-2 isolates that shows their relationship to one another can be found in Li et al. (End of February 2020). The isolates are divided into two main groups (L-type after the amino acid leucine and S-type after serine ), which gave rise to the assumption that the virus could have split into two (differently infectious) branches. However, in the opinion of other experts, at the beginning of March 2020 it was still too early to be able to make any clear statements about this. The in two main branches of the tree basal isolates lying derived from Wuhan (Hubei Province), which shows that, after current data, the virus has not start from there. Of course, this does not mean that there could have been unknown forerunners from elsewhere, such as the Chinese Yunnan , in animals or humans; The introduction of host animals to China cannot be ruled out either (see below → Origin and host range) . Another study at the beginning of April identified three strains A, B and C at this point. Strain A is most similar to the bat virus BatCoV / RaTG13 and appears to have spread worldwide from Wuhan, but strain B is predominant in mainland China itself and is also widespread in East Asia as well as in China. Tribe C is the main type in Europe.
The virus apparently mutates relatively slowly (one to two mutations per month), which means that it takes two to four times as much time as influenza viruses . This means, on the one hand, that genome analysis does not give a very high resolution of the path of spread of the virus; on the other hand, it gives hope that immunity acquired after surviving illness will last for a long time (for months). However, Icelandic virologists from deCODE Genetics ( Icelandic Íslensk erfðagreining ) had identified forty different mutations in infected people from this country alone by March 24, 2020. One of the affected persons was co- infected with two different forms of SARS-CoV-2 .
When monitoring genetic diversity and the development of the virus, a distinction must be made:
- Silent mutations thatdo not affect the encoded proteinsbecause of the degeneration of the genetic code anddefinea molecular genetic clock ,
- Mutations with effects on the phenotype (the appearance of the virus in all its forms). In the case of SARS-CoV-2, these apparently indicate ongoing adaptation to its new human host. For the development of antibodies and vaccines, it is important to find out which parts of the encoded proteins remain stable and are preserved so that the agents do not quickly become ineffective through adaptation of the viruses.
A detailed analysis of these facts can be found in Lucy van Dorp et al. (2020), see also Ärzteblatt of May 6, 2020: the infectiousness of the virus could increase over time, a fear that basically also applies to MERS-CoV, which has so far been difficult to transmit from person to person. A summary in German can be found on scinexx.de from May 8, 2020.
The dominant in the West form of the virus, which was strong in Europe spread from February 2020 and from there to other countries, has a mutation D614G in Spike - protein , thereby deviating from the Wuhan-variant from. In particular, this mutation increases the number of spikes on the surface of the virus particles by four to five times. As a matter of principle, such variants resulting from mutations must be taken into account when developing the vaccine.
Molecular Genetics and Phylogenetics

As usual in coronaviruses, the virus genome consists of single-stranded RNA ( ssRNA ) with positive polarity . The isolate Wuhan-Hu-1 (NCBI GenBank number MN908947) comprises 29,903 nt ( nucleotides ) with two 265 nt and 229 nt long untranslated regions at the 5 'end and at the 3' end, respectively. The putative (presumed) genes could code for ten proteins : a 7096 amino acid ( AA ) long ORF1ab polyprotein (replicase complex), a 1273 AA long surface glycoprotein (S for English spikes , compare Peplomer ), a 75 AA long coat protein (E for envelope , compare virus envelope ), a 222 AS long membrane glycoprotein (M), a 419 AS long nucleocapsid - phosphoprotein (N) and another five proteins (ORF3a, ORF6, ORF7a, ORF8 and ORF10). The sequence of genes corresponds to that of the SARS virus and that of all coronaviruses.
As of February 16, 2020, there were more than 40 complete genome analyzes of SARS-CoV-2 isolates. The genome size is between 29,825 and 29,903 nt. The GC content (the proportion of the nucleobases guanine and cytosine ) is 38.0 mol percent. The two virus isolates HKU-SZ-002a (NCBI-GenBank number MN938384) and HKU-SZ-005b (NCBI-GenBank number MN975262) come from patients in a family in Shenzhen and differ only in two nucleotides. The genomic analysis of these two isolates revealed that they closely related to the at bats (English bat ) occurring SARS-like coronaviruses bat-SL-CoVZXC21 (NCBI GenBank number MG772934) and bat-SL-CoVZC45 (NCBI GenBank number MG772933 ), there is a match in the nucleotide sequence of 89% for the latter. The genome of the two bat coronaviruses was sequenced in 2018, bat-SL-CoVZC45 was found in the Chinese horseshoe bat ( Rhinolophus sinicus ) from the family of the horseshoe bat (Rhinolophidae), the host animals were found in Zhoushan in the east Chinese province of Zhejiang in 2015 and Investigated in 2017.
Another virus isolate (WIV04, NCBI-GenBank number MN996528) of SARS-CoV-2 from the bronchoalveolar irrigation fluid of one of the first patients also shows the greatest phylogenetic similarity to one in another bat species ( Java horseshoe bat , scientifically Rhinolophus affinis , English intermediate horseshoe bat , common in Indonesia (Java), India, Vietnam, China) in the Chinese province of Yunnan, isolated coronavirus BatCoV RaTG13; the genome sequences are 96.2% identical. A genetic analysis published on January 27, 2020 also pointed to bats as the alleged original host of the virus. On January 29, 2020, the journal The Lancet published a genetic analysis of ten virus samples that had been obtained from nine patients. Accordingly, the genome sequence of all ten samples was 99.98 percent identical, which indicates that the newly discovered coronavirus variant only recently passed into humans. The genome sequence agrees to 88% and 87% percent with the genome sequences of bat-SL-CoVZC45 and bat-SL-CoVZXC21 which occur in bats. The ten samples, on the other hand, only show around 79 percent agreement in the genome sequence with SARS-CoV and around 50 percent with MERS-CoV . The results of the phylogenetic examinations are also illustrated as a phylogenetic tree , which shows the relationships of SARS-CoV-2 within the Coronaviridae . A presentation based on this can be found in the article Betacoronavirus .
The structure of the genome of both the SARS-CoV-2 isolates and the bat coronaviruses mentioned is typical for viruses of the Lineage B (sub-genus Sarbecovirus , English SARS-like Betacoronavirus ) of the genus Betacoronavirus . Due to the genetic distances to SARS-CoV and MERS-CoV, SARS-CoV-2 was initially seen as a new beta coronavirus species that infects humans . However, due to the great genetic similarity to the original SARS coronavirus , the Coronavirus Study Group of the ICTV proposed on February 11, 2020 that the new virus should be assigned to the same species as Severe acute respiratory syndrome-related coronavirus as the previous one.
The S-protein (S for English spikes ) is responsible for the binding to the host cell , functionally it is differentiated into the S1 domain and the S2 domain. The S1 domain mediates the binding to the surface receptor of the host cell, the S2 domain mediates the fusion of the cell membrane , and the virus then enters the cell through endocytosis . The S-gene of SARS-CoV-2 shows with 75% a rather low correspondence in the nucleotide sequence with the two bat isolates bat-SL-CoVZC45 and bat-SL-CoVZXC21 compared to the genome analysis. In particular, the nucleotide sequence that codes for the S1 domain differs significantly from these (68% agreement) but is more similar to the corresponding nucleotide sequence of BatCoV RaTG13. It has been shown that SARS-CoV-2 and SARS-CoV use the same cell receptor, the angiotensin-converting enzyme 2 (ACE2). This has been reliably proven experimentally, compare COVID-19 # Development of disease in COVID-19 .
morphology
The viruses that have been propagated in a cell culture over several days can be prepared for examination in a transmission electron microscope (TEM) after being separated by ultracentrifugation , using negative contrasting . The TEM image shows virions from spherical to pleomorphic shape with a diameter of 60 to 140 nanometers (nm). Spikes 9 to 12 nm long can be seen on the surface . The morphology corresponds to that of other known representatives of the Coronaviridae family. The host cells, which show a cytopathic effect in the light microscope image , can also be examined with the TEM after fixation and a subsequent ultra-thin section (thickness of 80 nm). In addition to virions, inclusion bodies that contain virus-filled membrane - bound vesicles in the cytoplasm are also found here.
Replication cycle
The virus replication cycle takes place in several stages.
First, the virions attach to the surface of the host cells. This happens specifically via certain surface features (receptors) of the host cell, in the case of SARS-COV-2 via the binding of the viral glycoprotein S to the ACE2 receptor . The host cells' ACE2 receptor could therefore be a possible starting point for therapy (1). However, it has not yet been sufficiently clarified whether other cell surface molecules can be used as binding partners for the glycoprotein S. Compared to SARS-CoV , the SARS-CoV-2 glycoprotein has developed an RGD peptide sequence, which means that receptors from the integrin family can also be used as binding partners.
After binding to the ACE2 receptor , the membrane-bound serine protease TMPRSS2 cleaves the viral glycoprotein S, whereby S is activated as a fusogenic protein and entry into the host cell occurs. TMPRSS2 is also a potential starting point for an effective drug (2).
In the next step, the pathogens penetrate the host cell (simplified representation) (3).
Before the virus starts to multiply , the genetic material ( RNA ) of the virus is released from the capsid (only one possible route shown) (4).
The actual process of reproduction, replication, now follows . Since SARS-COV-2 has RNA of positive polarity, the RNA can be used directly as “building instructions” (mRNA) for virus-specific proteins ( translation ). For the host cell, the viral RNA is practically indistinguishable from its own mRNA and the synthesis apparatus ( ribosomes ) of the host cell produces virus-specific proteins (S, M, E, N, RNA polymerase ) (5).
The genetic material (RNA) of the virus is copied in the host cell (RNA replication). The host cell's enzymes are unable to do this themselves; this task is taken over by the viral RNA polymerase, which makes many copies of the entire virus RNA (6).
If viral RNA copies and virus proteins are produced in sufficient quantities by the host cell, they are taken up in the endoplasmic reticulum (ER) and form new viruses (self-assembly) (7).
The finished virus particles are pinched off from the ER as Golgi vesicles (budding) (8).
The viruses emerge from the host cell by budding (9).
Origin and range of hosts
Since the disease was recognized as a viral disease, various groups of animals have been discussed as the origin or at least carrier of the pathogen. A molecular dating estimate based on genome comparison of the various SARS-CoV-2 isolates suggests that the virus variant originated in November 2019. Van Dorp and her colleagues determined on the basis of phylogenetic analyzes of the various virus variants in early May 2020 that the virus should have jumped to humans between October 6 and December 11, 2019. A detailed discussion of the various theories of ancestry - including controversially discussed - from July 2020 can be found in Sallard et al.
A comparative study on the infection risk of SARS-CoV-2 / COVID-19 was carried out in August 2020 by Joana Damas et al. submitted. According to this, the binding potential of the spike protein to the respective ACE2 receptor is greatest in primates (humans, bonobo , common chimpanzee , western lowland gorilla ), but very low in the following species: California sea lions , house mice , American crows and Mississippi alligators .
Snakes and birds
At the beginning of the epidemic in China, experts suspected that another mammal or poultry might be the main host . The transition from animals to humans could, however, have taken place via an as yet unidentified intermediate host . In the Journal of Medical Virology, Chinese researchers referred to snakes such as the multi-banded krait ( Bungarus multicinctus ) and the Chinese cobra ( Naja atra ), which are found on the wholesale market, which is believed to be the site of infection for the first infected, along with other living wild animals (so-called Ye Wei ) such as bats or rabbits . This hypothesis has been declared unlikely by other virologists, as there is currently no evidence that coronaviruses can also infect reptiles. So far, corona viruses have only been found in mammals and birds. Further research results on chickens and ducks ( Galloanserae ) s. u.
Pangolins and bats
Horseshoe bats - possibly several cave-dwelling species - were the reservoir of the pathogen SARS-CoV [-1], which triggered the SARS epidemic a few years earlier, with the larval roller ( Paguma larvata ) as a possible intermediate host between bats and humans. Since then, various other beta coronaviruses (in particular SARS-like ones of the subgenus Sarbecovirus ) have been found primarily in bats, but also in humans.
SARS-like coronaviruses (i.e. the subgenus Sarbecovirus ) were found in caves in the Chinese province of Yunnan in 2017 in various species of horseshoe bat : the Java horseshoe bat ( Rhinolophus affinis , English intermediate horseshoe bat ), the Chinese horseshoe bat ( R. sinicus ) and the great horseshoe bat ( R. ferrumequinum ); as well as in the Stoliczka trident leaf nose ( Aselliscus stoliczkanus ).
Due to the similarity of the binding site ( English receptor binding domain , RBD) of the spike protein to the human receptor ACE2, the virus isolate BatCoV RaTG13 (found in Java horseshoe bats Rhinolophus affinis , English intermediate horseshoe bat in Yunnan, fragments also applies to) sick and deceased miners from Yunnan 2016), as an important candidate for the origin of SARS-CoV-2, even if it is not clear whether the transmission took place directly. The match of the genome sequence between RaTG13 and SARS-CoV-2 is 96%.
After coronaviruses with a high genetic correlation to SARS-CoV-2 were found in Malay pangolin ( Manis javanica , English Sunda pangolin ) (Manis-CoV, more precisely Pan_SL-CoV_GD / P1L, isolates SRR10168377 and SRR10168378), these were suspected of origin of the pandemic, especially since these are traded in China despite the ban. The agreement in this case was 90% over the entire genome, but 99% in a specific region of the spike protein (S protein), which allows the virus to bind to the ACE receptors of human cells.
Interestingly, the RaTG13 virus isolated in the Java horseshoe bat R. affinis is comparatively different to SARS-CoV-2 in this genome section with only 77% agreement.
This means that the coronaviruses isolated from the Malay pangolins can penetrate human cells, but the one isolated from Java horseshoe bats cannot. In addition, this result is compatible with the assumption that the novel coronavirus SARS-CoV-2 could be the result of a recombination of the RNA molecules of two different viruses, one from the RaTG13 from bats from Yunnan, the other from the Pan_SL-CoV_GD from pangolins from Related to Guangdong. Then SARS-CoV-2 would have emerged as a new chimera of two viruses that were each very close to these two lines. This assumption was supported by another study by Xiaojun Li and colleagues (Duke University, Los Alamos National Laboratory, University of Texas, El Paso, and New York University) in late May 2020.
Coronaviruses, unlike influenza viruses, have an unsegmented genome (monopartite), i. H. only a single nucleic acid molecule (here RNA). The fact that there is still a recombination mechanism in this virus family has already been described earlier, in particular to explain the origin of the old SARS virus SARS-CoV [-1]. Such a recombination, regardless of whether the genome is segmented or unsegmented, can lead to a new virus that can infect a new host species and make some sick. The recombination event can therefore become the starting point for a new epidemic, as is suspected in SARS (and always feared in influenza). The prerequisite is the double infection (co-infection) of a (single) host individual by the two original viruses. However, so far (as of June 2, 2020) it remains unclear in which species the hypothetical double infection could have occurred and under what circumstances this could have happened.
As an alternative scenario that works without recombination, the following has been suggested on various occasions: The common ancestors of RaTG13 and SARS-CoV-2 originally come from the pangolin coronaviruses, from whose strain they are more similar to SARS-CoV-2 than 140 years separated. This line split up again about 40–70 years ago: one line remained in bats and lost the ability of its spike protein to bind to human ACE2. The other retained this ability and last jumped over to humans as SARS-CoV-2. The various possibilities are also discussed by Halloy et al. discussed in a PrePrint from July 2020. Boni et al. assume at the end of July 2020 that SARS-CoV-2 did not arise directly from a recombination of bat and pangolin coronaviruses, but that its line of development separated from that of the bat virus RaTG13 about 50 years ago.
Raccoon dogs as possible intermediate hosts
According to Christian Drosten could raccoon dogs ( Nyctereutes procyonoides , a Fuchssart) may be the desired intermediate hosts to be. The original SARS virus ( SARS-CoV-1 ) was also found in raccoon dogs, which are bred in China for their fur and can therefore be considered as a vector for humans.
Pets as hosts
According to the WHO, as of March 2020 there was no evidence that pets spread SARS-CoV-2 as a carrier. However, some other viruses from the corona virus family Coronaviridae can also cause diseases in domestic animals, e.g. As the two Alpha coronaviruses CCoV (dogs) and FCoV (cats).
dogs
On February 28, 2020, the Hong Kong government announced that it had tested positive for the virus for the first time in a dog living in the household of its infected owners. The WHO confirmed that the SARS-CoV-2 samples had been tested "weakly positive". Although the virus could be detected in the dog's blood, it did not trigger any clinically detectable evidence of disease in the dog. The animal was last tested for SARS-CoV-2 on March 12 and 13, 2020 with negative results, so its quarantine was ended and it was returned to the owner. Two days after the end of the quarantine, the dog died without any direct connection to the virus infection being detectable.
In mid-March, two more dogs in Hong Kong tested positive for SARS-CoV-2, which were also without any noticeable symptoms of infection.
In mid-April 2020 an article was published about the possibility of stray dogs as intermediate hosts for the transmission of Sarbecoviruses (RaTG13, Pangolin-CoV) from wild animals (bats, pangolins) to humans. The zinc finger protein ZAP plays an important role in this .
Cats
In Liège (Belgium) at the end of March 2020, an infected person's domestic cat tested positive for SARS-CoV-2. The animal developed diarrhea, had breathing problems and died. An infection in a domestic cat in Hong Kong at the end of March 2020 was, however, symptom-free. Antibody detection had previously shown in Wuhan that cats had also been infected there. In addition, it has been proven several times in laboratory experiments that infected cats can pass the virus on to other cats. There is a suspicion that a cat could have transmitted the virus between residents of a nursing home in Bavaria, although they were isolated from one another.
More vertebrates
In early April 2020, an adult tiger in New York's Bronx Zoo tested positive for SARS-CoV-2 after being noticed a dry cough and wheezing, but no shortness of breath. Two lions and five tigers also had similar symptoms, which is why they were also suspected of being infected with SARS-CoV-2. The animals were probably infected by an asymptomatic employee of the zoo. The animals recovered a few days after the appearance of symptoms. At the Joburg Zoo in Johannesburg ( South Africa ), a puma was infected by an infected animal carer in July 2020 .
Laboratory experiments in Korea have shown that ferrets are susceptible to SARS-CoV-2 infection and can pass it on to conspecifics. The Friedrich Loeffler Institute confirmed the findings from Korea based on its own tests and at the same time pointed out that Egyptian bats are also susceptible to SARS-CoV-2 infection, but pigs and chickens are not. The susceptibility of ferrets in particular is an important finding, "since they could be used as model animals for the infection of humans to test vaccines or drugs". Chinese researchers reported in the journal Science in April 2020 that the virus reproduced poorly in dogs, pigs, chickens and ducks, and confirmed that ferrets and cats can be infected. Even hamsters that had developed [-1] only very mild symptoms after infection with SARS-CoV and therefore as model animals were unsuitable, could be infected in the laboratory with SARS-CoV-2 showed clear symptoms and had high levels of virus in the lungs and Bowel on. For more research results on fruit bats, see p. u.
In April and May 2020, infections and diseases were found in several Dutch mink farms. The virus was probably brought in by infected employees and then passed on from animal to animal. The diseased mink showed symptoms similar to humans: respiratory problems, problems with the digestive tract, increased mortality. Viral RNA was detected in the air in the animal husbandry, which was polluted by fine dust . Detailed analyzes of the genetic code of the SARS-CoV-2 variants circulating in the farms and in the vicinity of the farms provided evidence that two infected employees of the farms were infected with the mink and that several cats roaming freely in the area of the farms were also infected "Farm-typical" SARS-CoV-2 infections, which is why they are also possible carriers of viruses to the mink.
These results were supported by a study by Kore Schlottau ( WHO ) et al. (published in July 2020) confirmed and deepened once more. Were tested Egyptian fruit bats ( Rousettus aegyptiacus , English fuit bats ), ferret (by the authors as Mustela putorius called), pigs ( Sus scrofa domesticus ) and domestic fowl ( Gallus gallus domesticus ). The domestic pigs and domestic chickens also turned out to be not susceptible to SARS-CoV-2. Seven out of nine Egyptian bats first developed rhinitis , and as the disease progressed, the virus migrated through the trachea and sometimes into the lungs. In the ferrets, virus replication was even more efficient, but no symptoms except for possible mild rhinitis. Like the Egyptian bats, they developed antibodies against SARS-CoV-2.
While infected mice apparently developed no symptoms of the disease in the laboratory, Y.-C. Wang and colleagues in China were able to use CRISPR / Cas9 to replace the ACE2 of the mice (mACE2, murine ACE2) with that of humans (hACE2, human ACE2) in C57BL / 6 laboratory mice . The hACE2 mice showed virus replication of SARS-CoV-2 in their lungs, trachea and brain. The digestive tract was also affected, as seen in some human patients. They seem suitable, for example, to test a vaccine before it is given to humans; an alternative to the method of testing the effect of an agent on artificially mutated Sarbecoviruses, as happened recently with Remdesivir and SARS-CoV / SARS-CoV-2 RdRp (old SARS virus with RdRP gene from SARS-CoV-2).
Primates
Whether next to people or apes can fall ill with SARS-CoV-2, is currently (as of end of March 2020) still unsettled.
- In 2016 was in chimpanzees in Tai National Park ( Ivory Coast an infection with the human coronavirus OC43 (HCoV-OC43, a) beta coronavirus from the subgenus Embecovirus , species Beta coronavirus 1 ) is observed, causing the mild in humans cold-like symptoms. These were also shown by the chimpanzees. It cannot therefore be ruled out that SARS-CoV-2 could at least jump over from humans to great apes. It was recommended (especially for game rangers) to keep a minimum distance of 7 to 10 meters from the wild animals and to adhere to appropriate quarantine times for the animals.
- In addition to chimpanzees (including bonobos ), gorillas and orangutans could also be threatened, such as chimpanzees and gorillas from the human metapneumovirus (HMPV, family Pneumoviridae ).
- A Chinese research group led by Chuan Qin made the preliminary results of their studies on rhesus monkeys available as a preprint in March 2020 . It was particularly about the question of infectivity after surviving illness. A study on rhesus monkeys published in Science in May 2020 also reported "protective immunity" after the first illness.
- Dutch researchers reported in March 2020 Science that SARS-CoV-2 in cynomolgus monkeys, a "COVID-19-like illness" were causing, which is why these animals as a model for testing preventive and therapeutic strategies appropriate.
Risk group according to the Biological Agents Ordinance
In Germany, the Biological Agents Ordinance (BioStoffV) applies to employees who may come into contact with infectious agents through their work . The Committee for Biological Agents (ABAS ) set up at the Federal Institute for Occupational Safety and Health (BAuA) temporarily assigned SARS-CoV-2 to risk group 3 according to the BioStoffV (second highest level) on February 19, 2020 . Basically, the classification into risk groups takes place in the technical rules for biological agents (TRBA), which are published by the BAuA, for viruses this is the TRBA 462: Classification of viruses in risk groups . If new, not yet assigned pathogens occur, a preliminary classification is carried out by the ABAS. In the justification, reference is made to the similarity of SARS-CoV-2 with the SARS-CoV-1, which triggered the SARS pandemic in 2002/2003, and the similarity to a lesser extent with the MERS-CoV is also mentioned. These two viruses were also assigned to risk group 3. The ABAS cites the "currently lacking options for vaccination prevention and therapy as well as the great possibility of dissemination in the population" as the reason for the provisional assignment to risk group 3.
In addition, recommendations are given for working with the virus during diagnostics in the laboratory: Non-targeted activities (see Section 5 BioStoffV ) - based on the test material, e.g. sample preparation, sample preparation and inactivation, in order to provide detection using RT-PCR (see section Detection methods ) - can be carried out under the conditions of protection level 2. All activities in which aerosol formation is to be expected must be carried out in a class II microbiological safety workbench . In addition, the appropriate personal protective equipment must be worn. Specific activities according to § 5 BioStoffV may only be carried out in laboratories of protection level 3. B. the multiplication of the virus in a cell culture . The American health authority CDC had previously issued similar recommendations.
Clinical manifestations
| Classification according to ICD-10 | |
|---|---|
| U07.1 | COVID-19, virus detected |
| U07.2 | COVID-19, virus not detected |
| ICD-10 online (WHO version 2019) | |
Detection methods
Proof of procedure
- → See: COVID-19 # Diagnostics
RT-PCR test
The Charité's detection method is the real-time quantitative reverse transcriptase polymerase chain reaction (abbreviated as qRT-PCR or RT-qPCR).
The test reacts to the presence of two specific short gene sequences ( nucleotide sequences ), which are characteristic of the viruses mentioned, referred to as the E gene and the RdRp gene. The E gene codes for the virus envelope ( E for envelope ), the RdRp gene for the RNA-dependent RNA polymerase ( RdRp for RNA-dependent RNA polymerase). The test therefore also reacts to virus residues.
The PCR test is considered the gold standard for the detection of the new coronavirus SARS-CoV-2.
procedure
The test includes three standardized individual tests ( called " assays " in German ) in a defined process sequence:
- Screening: The first assay (E-gene) serves as a screening because it detects several virus species of the subgenus Sarbecovirus (from the genus Betacoronavirus ).
- Confirmation: If the screening is positive, a confirmation assay (RdRp gene) must be performed.
- Characterization: If a positive result is also obtained there, a third characterization assay (also RdRp gene) follows.
The last two assays use (among others) a gene probe that is specific for SARS- CoV-2 . The second assay also contains a probe that matches the nucleotide sequence of both the RNA part of SARS- CoV-1 and SARS- CoV-2 .
The qRT-PCR takes 90 minutes to three hours. Instead of the real-time quantitative variant, the products of reverse transcriptase PCR can also be detected using agarose gel electrophoresis .
development
The first version of the real-time RT-PCR assay was created before the genome sequence of SARS- CoV-2 was published. For the primer design , the first SARS-associated coronavirus SARS-CoV -1 and other SARS-associated coronaviruses ( Sarbecoviruses ) that occur in bat species were used. After the genome sequence was published, the primers were selected that are suitable for the detection of SARS- CoV-2 .
The molecular biological detection method, which is used in the Consiliarlabor at the Charité, was developed in collaboration with the Berlin biotechnology company TIB Molbiol , a first version was already available on January 13, 2020. Chinese scientists have willingly contributed unpublished results to this. Basic data came from international research networks, and the global section of the European Virus Archives (EVAg) supplied the necessary products (SARS- CoV- 1 RNA and RNA transcripts ) for the assays. Other groups of scientists have also published the methods they have developed. These are PCR protocols or lists of suitable primers and their substance concentration used for RT-PCR , for example from the Centers for Disease Control and Prevention (CDC) in the USA, the CDC in China or the University of Hong Kong . They differ in which genes of the virus RNA are detected, for example the N gene ( N for the nucleocapsid phosphoprotein), the ORF1ab gene (codes for the ORF1ab polyprotein) or the gene for the spike protein. The detection method developed in Berlin has already sold 40,000 test kits for examining 4 million samples in over 60 countries in the first two months.
In Germany, a method was presented in March 2020 in which samples from several test persons are combined and tested together. A negative result means that the results of all samples are reliably negative, and only if the result is positive do the samples need to be examined individually. Therefore, according to the researchers, significantly more people can be tested with comparable effort than with conventional methods, provided that the people examined have a low likelihood of infection. According to the researchers, this could increase the number of daily examinations in Germany to between 200,000 and 400,000.
Expressiveness
The most important quality criteria for the validity of diagnostic laboratory tests are specificity and sensitivity .
Under specificity , the probability is understood that a really negative sample is recognized as negative (excluding false positives). Under sensitivity, on the other hand, the probability that an actually positive sample will also be recognized as positive (exclusion of false negatives).
If the prevalence is low and the test indication is low (e.g. testing of asymptomatic persons), high demands are placed on the specificity of the test.
Specificity
The test reacts positively to SARS- CoV-2 and SARS- CoV-1 . The cross-reactivity to SARS- CoV-1 is intentional in order to make SARS- CoV-1 (from laboratory stocks) available as a positive test control. Further data from the test development indicate that the test for "probably all Asian viruses" (translated from English) from the subgenus Sarbecovirus gives a positive result.
During development, it was ensured that the test does not react positively to the endemic human coronaviruses ( HCoV-229E , -NL63 , -OC43 , -HKU1 ), the MERS-CoV and many other common pathogens that cause respiratory diseases. Like other established tests for human coronaviruses, this test also reacts positively to various coronaviruses unknown in humans (especially those from certain bat species).
Samples that do not contain the target virus are tested to determine that the test does not react unintentionally positive to another virus . This ensures that the test actually shows a negative sample as negative in these cases. On the basis of these investigations, the specificity of this test is estimated to be extremely high, provided that (for other reasons) it can be ensured that the tested sample is free from SARS- CoV -1 and other Asian Sarbecoviruses.
An example from 2006 of a PCR test for SARS- CoV-1, which is closely related to SARS - CoV-2 , shows that even long-established tests can in retrospect show deficiencies in specificity . It was shown that this SARS- CoV- 1 test also reacted positively to the genus-related human beta coronavirus HCoV-OC43 . Here, parallel antibody tests initially supported the false positive result. This means that this cross-reaction could also occur in other SARS- CoV -1 tests.
Although the SARS- CoV-2 test described in this section also represents a SARS- CoV-1 test at the same time , this specific cross-reaction is evidently excluded here because, as mentioned above, it has also been specifically validated against the HCoV-OC43 . Mutations on the HCoV-OC43 could change that again in the future.
In a collaborative study of the German maintenance e. V. (Society for the Promotion of Quality Assurance in Medical Laboratories), the quality of 463 laboratories from 36 countries was examined in May and June 2020, among other things, to determine whether they can reliably rule out false positive results in the tests. The laboratories achieved mostly correct negative results for the SARS-CoV-2 negative samples (97.8% to 98.6%). Laboratories that could not only deliver 100% correct results were not certified.
In June 2020, the Foundation for Innovative New Diagnostics in Geneva determined the sensitivity and specificity of SARS-CoV-2 test systems from 21 manufacturers by comparing them with their own in-house test. The specificity was determined on the basis of 100 negative samples and had a range from 96% to 100%. So far it is unclear whether the results interpreted as false-positive are actually false-positive or can be traced back to false-negative reference results of one's own in-house test.
In a publication from the Friedrich Loeffler Institute Pitfalls in SARS-CoV-2 PCR Diagnostics , the authors report on the occurrence of manufacturer-side contamination of commercial primer / probe sets with the SARS-CoV-2 target sequence of the RT-qPCR. Since contamination of the reagents used can also lead to incorrect test results, the authors recommend batch testing and systematic quality controls during the test process.
sensitivity
While the high specificity has been widely accepted, the sensitivity of the test has been criticized and frequent false negative results are reported. As a result, the z. For example, in patients tested several times in a row, the status kept changing between positive, negative and unclear results. The problem is not seen here on the "technical" side of the test, but in the correct execution and handling.
There may be too few samples, samples from the wrong places, or samples taken in the wrong way. This can result in the virus being missing from the sample but still present in humans. The test then turns out to be "technically correct" negative, even though the person carries traces of the virus.
The above-mentioned systematic review of the tests of various manufacturers by the Foundation for Innovative New Diagnostics in Geneva in June 2020, in which the sensitivity was determined on the basis of 50 positive samples in each case, showed that the sensitivity ranges from 90.00% to 100.00%.
Prevalence
The prevalence , also known as the pretest probability or pretest probability, is a factor that has a significant influence on the rate of false-positive and false-negative results, but is neither caused by the care taken in collecting and transporting the sample, nor by the care taken during use of the test kit or its quality in terms of sensitivity or specificity. The effect of different prevalence values on the rate of false-positive and false-negative results in the context of the PCR tests for SARS-CoV-2 can be illustrated interactively: If COVID-19 hardly occurs in the group tested, the probability of it decreases If the test result is positive, the rate of false-positive tests increases if you are actually infected. This fact is misused by deniers of the corona pandemic, but at the same time it is one of the reasons for the repeated recommendation from politics and the laboratory association that PCR tests should not be used “without cause”, but targeted.
Other methods
Genome analysis
Laboratories equipped for genome analysis ( DNA sequencing of the genome ), i.e. an automated sequencing system , can also identify SARS-CoV-2 in this way. Complete genome analyzes of SARS-CoV-2 isolates for comparison are available, for example, in the gene database of the National Center for Biotechnology Information (NCBI) or via the GISAID platform (see section Molecular Genetics).
Nucleic acid detection

The World Health Organization (WHO) reported in mid-January 2020 on the development of simplified molecular biological methods, the Nucleic Acid Amplification Technology (NAAT), the assays of which have been validated. The NAAT method is also based on RT-PCR, but the ready-made assay has the advantage of being easier to use and can be used by appropriately equipped routine laboratories.
On February 5, 2020, the US American authority CDC announced that it would make such an assay (test kit) available for use in accredited diagnostic laboratories . The assay is called Centers for Disease Control and Prevention (CDC) 2019-Novel Coronavirus (2019-nCoV) Real-Time Reverse Transcriptase (RT) -PCR Diagnostic Panel and is used for the detection of both the novel coronavirus and SARS-like coronaviruses provided in samples of the upper and lower respiratory tract of patients. Prior to this, the Food and Drug Administration (FDA) granted accelerated approval , which means that since February 4, 2020, the assay can also be used outside research institutions. A test kit enables 700 to 800 samples to be examined, 100 of these packs went to US laboratories, and a further 100 to international laboratories that carry out tests on behalf of the WHO, for example. The examination takes about four hours from the preparation of the sample to the availability of the results. The Hamburg company Altona Diagnostics and the Darmstadt company R-Biopharm have been offering a similar test kit for RT-PCR since February 2020, which is also sold worldwide. A test kit enables the analysis of around 100 samples. However, so far these have only been approved for use in research.
Rapid test
The term rapid test used in the media is not clearly defined; on the one hand, it is associated with the expectation that the time until a test result is available compared to the RT-qPCR method, which is now routinely used, on the other hand, it can also mean that it is It is a point-of-care-testing (German near- patient laboratory diagnostics ) in which the step “sample transport for analysis in the central laboratory” is omitted, since the corresponding devices are used on site, for example in a doctor's office. Thus, cartridge systems based on the RT-PCR method are sometimes referred to as rapid tests in the media, but are presented in a separate section here.
At the end of January 2020, Xinhua reported that the Chinese National Medical Products Administration (NMPA) approved four test kits for a new test procedure on January 26. The assay is from the biotechnology company Sansure Biotech from Changsha been developed. With the help of suitable laboratory automation , test results should be available after just 30 minutes. The state news agency said that a company based in Wuxi , eastern Jiangsu Province , worked with the National Institute for Viral Disease Control and Prevention to develop a rapid method. With the test kit, the virus should be detected within 8-15 minutes, the company can produce so many test kits every day that it is possible to examine 4,000 samples and the method is said to have already been used in the province of Hubei. The Xinhua reports contain no reference to the molecular biological methods used. Furthermore, at the Chinese Tianjin University, in cooperation with a Beijing biotechnology company, a rapid process is being developed that should produce results after 15 minutes. It is in trial use (as of February 2020).
Researchers at the Hong Kong University of Science and Technology announced the development of a portable device with which the novel coronavirus should be detectable within 40 minutes in early February 2020. Modified chip thermal cyclers are used for the faster process compared to conventional qRT-PCR . Researchers from the Institute for Health Innovation & Technology (iHealthtech) at the National University of Singapore also reported on developing a rapid method in February 2020. It is based on the enVision technology, which has been used since 2018 , with which nucleic acids can be detected within 30 to 60 minutes. It is estimated that several months will be needed before the new test procedure is ready for the market .
A German research group at the University of Bonn led by Hendrick Streeck presented the results of a rapid test validation in April 2020. The CoV-2 Rapid Test was tested as part of a population screening and compared with samples obtained in parallel for PCR diagnostics. Of a total of 49 people, 22 were positive in the PCR; however, the rapid test only correctly identified eight of them as positive (sensitivity 36.4%). Of the 27 PCR-negative persons, 24 were correctly diagnosed as negative by the rapid test (specificity 88.9%).
Cartridge test
In a cartridge system, a technically elaborate, but still transportable by the size dimensions of appliance is (engl. Analyzer ) is used, in which a designed for this cartridge (engl. Cartridge ) is used. The cartridge is previously filled with the sample material, e.g. B. equipped with the swab , additional chemicals and biological agents for sample preparation and analysis are contained in the cartridge. The cartridge, which is designed for single use, shortens the time until the test result is available and offers the user the advantage of minimizing contact with the infectious agents (see the section on risk group according to the Biological Agents Ordinance ). Cartridge tests, also as engl. panel , have been used since 2018 for the diagnosis of pathogens that cause respiratory diseases. Here, too, the method is based on RT-qPCR, the real-time quantitative reverse transcriptase polymerase chain reaction, but since the genes of several pathogens are analyzed at the same time, it is assigned to the multiplex PCR .
The biotechnology company Qiagen NV developed a cartridge test as a rapid test at its location in Hilden , which is based on a procedure that has already been approved internationally for the diagnosis of pathogens . a. Have SARS-associated viruses and EHEC detected. The procedure was expanded to include the detection of the genes ORF1b and E present in the SARS-CoV-2 genome and the results were compared with those of the RT-PCR method. The company worked with WHO to achieve validation . The portable diagnostic device is suitable for use in medical practices or at airports. A swab from the throat or a blood sample is suitable as sample material; test results are available within 60 minutes. In Germany, preliminary approval has been applied for by the Federal Institute for Drugs and Medical Devices . The diagnostic devices were tested in French and Chinese hospitals in February 2020 and received approval from the US and European authorities (as of March 27, 2020). Compared to the routinely used RT-qPCR method, cartridge tests are more expensive, and the associated analysis device is required.
Two other diagnostics companies in Germany are also developing such rapid tests. The US company Cepheid Inc received accelerated approval from the FDA in March 2020. This is also a cartridge system based on RT-qPCR with the associated analyzer , test results should be available after 45 minutes. Around 23,000 of the appropriate analysis devices are currently in use worldwide, mostly in hospitals; an analyzer can accommodate up to four cartridges at the same time (as of March 27, 2020). The Robert Bosch GmbH developed together with the British company Randox Laboratories also a cartridge test, should be available with the test results after 2.5 hours. The test system is scheduled to come onto the market in April 2020, but so far has only been approved for research institutions.
Antibody detection

The World Health Organization (WHO) reported in mid-January 2020 on the development of antibody detection as a serological test. This will result in an assay, for example an immunassay such as ELISA or a lateral flow test , with which antibodies from patient samples ( blood serum ) can be detected by an antigen-antibody reaction . The lateral flow test can be referred to as a rapid test (see above) in the sense of point-of-care testing (German: near-patient laboratory diagnostics ), a well-known example based on it is the pregnancy test .
Depending on which antibody is tested for, a current infection can still be detected (early antibody immunoglobulin M (IgM)) or an infection that has already been completed by late antibody immunoglobulin G (IgG); this antibody class change is known as seroconversion . The suggestion was made to use IgM antibody detection to diagnose acute infections, but the examination by Chinese scientists of 535 plasma samples from 173 patients showed that IgM was only detectable in just under 30% of the patients in the period 1 to 7 days after the onset of symptoms. while 73% of the patients over the 8-14 day period. Thus, the IgM antibody detection can only supplement the PCR testing.
The WHO and the Robert Koch Institute in Germany call to collect serum samples from confirmed or suspected cases in the acute phase and asservieren . The WHO recommends taking the first sample in the first week of illness and the second sample three to four weeks later. This can be used to check seroconversion. After contact with SARS-CoV-2, the antibodies immunoglobulin A (IgA) are formed after about three weeks (corresponds to about two weeks after the onset of symptoms ), and the IgG antibodies after about 4 weeks. When examining 153 patients, it was found that seroconversion takes place 20–30 days after the onset of symptoms, ie that IgG antibodies are present in sufficient quantities. Antibody tests for IgA and IgG are therefore not intended for the acute diagnosis of sick patients and are not a substitute for PCR analysis. Their results provide epidemiological data that can be used to determine the extent of the outbreak and help verify the effectiveness of vaccines .
The choice of the correct antigen is decisive for the informative value of the antibody test . If a particular antibody binds to more than one antigen, it is a cross-reactivity which leads to false positive test results because more than the actual antigen reacts. The structures described in the Characteristics section are suitable as antigens for SARS-CoV-2 , for example the nucleocapsid protein (N) or the spike protein (S) as a whole or, alternatively, the S1 and S2 domains. Possible cross-reactions to the coronaviruses SARS-CoV-1 , MERS-CoV , HCoV-HKU1 , HCoV-OC43 , HCoV-NL63 , HCoV-229E as well as coronaviruses occurring in cats and pigs are mentioned in the literature . When validating the developed antibody detection methods, diagnostic sensitivity (true positive rate) and specificity (true negative rate) are important quality criteria, which are illustrated with the help of the following table.
| sick (infected) | healthy (not infected) | |
|---|---|---|
| Test positive | positive (P) | false positive (FP) |
| Test negative | false negative (FN) | negative (N) |
The sensitivity of the test procedure indicates the proportion of those who tested positive to the total of those actually infected, while the specificity indicates the proportion of those who tested negative to the total of those who were actually not infected, expressed as formulas:
- or.
For a reliable test result, values close to 100% are aimed for for both criteria, a high sensitivity ensures that no infected person is accidentally overlooked, a high specificity indicates that no “false alarms” (e.g. due to cross-reactivity) are triggered. For the method validation of serological evidence, the reference method (also known as the gold standard ) is the neutralization test (NT), in detail the plaque reduction neutralization test (PRNT). For the patient sera used as samples, the SARS-CoV-2 has previously been detected in the patients by PCR or the other coronaviruses or other viruses in the controls. For the validity of the specificity, it is also important that samples with antibodies against many different viruses are tested.
The first ELISA tests were carried out in a Chinese research laboratory in January 2020, using the nucleocapsid protein (N) of a bat coronavirus similar to SARS-CoV-2 as the antigen. This enabled the antibodies IgG and IgM to be detected in serum samples from a patient and their titers to be determined over several days during the course of the disease. In a second test, serum samples taken 20 days after the first symptoms were examined. All patient sera, but not the sera from healthy individuals, showed a strongly positive IgG reaction, some patient sera also showed an IgM reaction, which indicates a current immune response, i.e. a current infection.
Commercial antibody detection tests have been available since March 2020. As part of the validation, two ELISA tests were checked that are based on an IgA and IgG reaction to the S1 protein; a rather low specificity was found, as there was cross-reactivity with the human coronavirus OC43 and thus false positives Results. The IgA ELISA performed better in terms of sensitivity. However, the validation study is only based on a small number of patient sera, on the one hand n = 10 of three COVID-19 patients, on the other hand n = 31 of nine COVID-19 patients.
Cell culture
The virus has been able to reproduce in a cell culture for research purposes in China , Australia , France , Germany and the USA , among others . The Chinese scientists use epithelial cells of the human respiratory tract , which simulate the multilayered mucociliary epithelial tissue ( ciliated epithelium ), and the Vero E6 and Huh-7 cell lines (isolated from human liver carcinoma ) are also used.
treatment
So far, there is no specific treatment for the disease COVID-19 ; therapy aims to alleviate the symptoms. However, it is being investigated whether known antivirals are also effective in the event of an infection with SARS-CoV-2.
prevention
Vaccine development
Immediately after the virus's RNA sequence was published, vaccine development began in several laboratories . The international vaccine initiative CEPI (Coalition for Epidemic Preparedness Innovations) planned to carry out the first tests with vaccines developed by then by mid-June 2020 . Several potentially suitable companies received financial support for this. In Germany this affected a. the Tübingen biotechnology company Curevac , which worked together with the Paul Ehrlich Institute on the rapid development of vaccines. The Robert Koch Institute pointed out that clinical studies with vaccines against MERS-CoV are currently ongoing. However, clinical studies are only the first step; if the study is successful, a vaccine would probably not be available for several months at the earliest, although it certainly could not be made available to the entire population in a first phase.
Vaccination can, however, lead to the development of non-neutralizing antibodies which may even facilitate the infection of the macrophages and thus worsen the infection. This has been observed, for example, when cats are vaccinated against feline coronavirus ; there, this increase in infection occurs not only because of a vaccination but also because of a previous illness with the virus.
Vaccination against other infections
The Berlin Senate Health Administration recommended the end of February 2020 all people over 60 and the chronically ill, their vaccination status to check and possibly vaccination against pneumococcal (vaccines as Pneumovax 23 were, however, in March 2020 only limited available) and whooping cough perform (pertussis) or refresh to to let. Since people over 60 years of age and the chronically ill are particularly at risk from SARS-CoV, they should be protected as a precaution.
Hygiene measures
The most important of these measures are:
- Personal hand hygiene (regular hand washing with soap for at least 20 seconds)
- Do not touch your eyes, nose or mouth with unwashed hands
- Keeping the minimum distance (1.5 to 2 meters) to other people except those in the same household
- Cough or sneeze only in a handkerchief or the crook of your arm, never in your hand
- Wearing mouth and nose protection in public transport, in hospitals, homes and z. T. also in public
- Ventilate closed rooms sufficiently and frequently
- If you feel sick, call the information phone instead of going to the doctor and stay at home
- Install one of the newly developed Corona apps on your mobile phone.
Epidemic situation
SARS-CoV-2 causes the novel disease COVID-19 (for English corona virus disease 2019 ), which became conspicuous in December 2019 in the megacity of Wuhan in the Chinese province of Hubei , and in January 2020 it developed into an epidemic in the People's Republic of China and also developed spread worldwide as the COVID-19 pandemic . In order to counteract the spread to states without efficient health systems, the World Health Organization (WHO) declared an international health emergency on January 30, 2020 . On March 11, 2020, the WHO upgraded the previous epidemic to a pandemic .
Reporting requirement
In Germany, direct and indirect evidence of the novel coronavirus has been reported by name since 23 May 2020 in accordance with Section 7 (1) No. 44a of the Infection Protection Act (IfSG) for laboratories , provided that the evidence indicates an acute infection. The obligation to notify was introduced by ordinance on February 1, 2020 . Since the statutory regulation by the Second Act for the Protection of the Population in the Event of an Epidemic Situation of National Impact in the IfSG, the test results (including negative test results) are not to be reported by laboratories by name (Section 7 (4) No. 1 IfSG). However, this non-named reporting requirement for test results (and thus for negative test results) is suspended as long as the Robert Koch Institute does not yet have the German Electronic Reporting and Information System for Infection Protection (DEMIS). In addition, doctors are obliged to report the respiratory disease COVID-19 caused by the virus .
In Austria there is also an obligation to notify , according to the Epidemic Act 1950 together with a regulation. The duty to report exists for suspected illnesses and deaths due to this virus. In addition, the segregation regulation was expanded to include the new corona virus.
There is also a reporting requirement in Switzerland. This follows from the epidemic law of Switzerland in connection with the epidemic Regulation and Regulation of EDI on the reporting of observations of communicable diseases of man. According to Appendix 1 of the EDI Ordinance, doctors must report a clinical suspicion and the initiation of a pathogen-specific laboratory diagnosis and the necessary epidemiological connection. According to Annex 3 of the EDI Ordinance, laboratories must report positive and negative findings (i.e. evidence). The Federal Office of Public Health has published criteria for suspicion, sampling and reporting.
Web links
- COVID-19 (Coronavirus SARS-CoV-2). Overview page of the Robert Koch Institute (RKI). In: rki.de. Robert Koch Institute, March 19, 2020(will be updated continuously).
- Answers to frequently asked questions about the SARS-CoV-2 coronavirus. In: rki.de. Robert Koch Institute, March 18, 2020(will be updated continuously).
- Coronavirus pandemic in Germany: challenges and options for intervention. (PDF; 158 kB) National Academy of Sciences Leopoldina , March 21, 2020.
- Answers to frequently asked questions about the novel coronavirus (SARS-CoV-2). In: infektionsschutz.de. Federal Center for Health Education , February 28, 2020(updated every week).
- Can the novel coronavirus be transmitted through food and toys? Updated questions and answers of BfR of 27 May 2020. In: bfr. Bund.de . Federal Institute for Risk Assessment , May 27, 2020.
- Current information on the coronavirus. In: bundesgesundheitsministerium.de. Federal Ministry of Health (Germany) , February 29, 2020(will be updated continuously).
- Information about the coronavirus. In: Sozialministerium.at. Federal Ministry for Social Affairs, Health, Care and Consumer Protection (Austria), March 6, 2020(is continuously updated).
- Novel coronavirus (COVID-19). In: Sozialministerium.at, Infectious Diseases AZ. Federal Ministry for Social Affairs, Health, Care and Consumer Protection (Austria), March 6, 2020(is continuously updated).
- New coronavirus. In: bag. admin.ch . Federal Office of Public Health (Switzerland), March 13, 2020(will be updated continuously).
- Q&A on COVID-19. In: ecdc.europa.eu. European Center for Disease Prevention and Control , February 16, 2020, accessed February 29, 2020 .
- World Health Organization : Coronavirus disease (COVID-19) outbreak. In: who.int. (English, is continuously updated).
- Terra X : The new coronavirus scientifically tested. In: zdf.de . January 30, 2020 .
- Harald Lesch : Corona: What does science know? Article in the science magazine Leschs Kosmos . In: ZDF website . March 24, 2020 (43 min, available until March 17, 2025).
- The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team: Vital Surveillances: The Epidemiological Characteristics of an Outbreak of 2019 Novel Coronavirus Diseases (COVID-19) - China, 2020. February 21, 2020, accessed March 1, 2020 .
- Largest official collection of expert answers for risk groups In: Austrian online platform for the chronically ill , March 30, 2020, accessed on March 30, 2020.
- Podcast with Christian Drosten on NDR , or the scripts for the broadcast in PDF
- WDR : Corona - the most important facts. In: Quarks Extra . April 4, 2020, accessed on March 28, 2020 (The time that the contaminated aerosol spends floating in the air before it sinks to the ground is given here with a maximum of 10 minutes, which does not correspond to the latest scientific status where longer times are mentioned , otherwise the article is up-to-date, very informative and well prepared to bring the topic closer to non-medical professionals).
- Zita Aretz, Victor Nicolaus: How the coronavirus infects our cells. In: n-tv. April 5, 2020, accessed on April 6, 2020 (The title refers explicitly to SARS-CoV-2. Illustrated with numerous schematic drawings, including hygiene measures to avoid infection and how it works).
Individual evidence
- ↑ Alissa Eckert, MS, Dan Higgins, MAM: ID #: 23312 . In: Centers for Disease Control and Prevention (Ed.): Public Health Image Library ( PHIL ). 2020, accessed February 26, 2020.
- ↑ a b c d ICTV: ICTV Taxonomy history: Severe acute respiratory syndrome-related coronavirus , EC 51, Berlin, July 2019; Email ratification March 2020 (MSL # 35)
- ↑ a b c d e f g Alexander E. Gorbalenya, Susan C. Baker, Ralph S. Baric, Raoul J. de Groot, Christian Drosten, Anastasia A. Gulyaeva, Bart L. Haagmans, Chris Lauber, Andrey M Leontovich, Benjamin W. Neuman, Dmitry Penzar, Stanley Perlman, Leo LM Poon, Dmitry Samborskiy, Igor A. Sidorov, Isabel Sola, John Ziebuhr: Severe acute respiratory syndrome-related coronavirus: The species and its viruses - a statement of the Coronavirus Study Group . In: bioRxiv . February 11, 2020, bioRxiv : 10.1101 / 2020.02.07.937862v1 ( preprint full text), p. 1–20 , doi : 10.1101 / 2020.02.07.937862 (English).
- ↑ National Institute of Allergy and Infectious Diseases: Novel Coronavirus 2019 . In: flickr . Retrieved March 18, 2020.
- ↑ Sebastian Thaler, et al .: Importance of corneal organ culture in donors with possible SARS-CoV-2 infection . In: The ophthalmologist . 2020, doi : 10.1007 / s00347-020-01152-z .
- ↑ Information on the detection, diagnosis and therapy of COVID-19 patients (PDF). Ed .: STAKOB office at the Robert Koch Institute , as of April 17, 2020, accessed on April 30, 2020.
- ↑ a b Novel Coronavirus (2019-nCoV). (PDF; 1.0 MB) Situation Report - 22nd WHO , February 11, 2020, accessed on February 13, 2020 .
- ^ A b Pneumonia of unknown cause - China. Disease Outbreak News (DONs) - WHO , January 5, 2020, accessed April 29, 2020 .
- ↑ Florian Rötzer: WHO calls out international emergency January 30, 2020.
- ↑ Coronavirus SARS-CoV-2: Risk assessment for COVID-19. “The worldwide spread of COVID-19 was declared a pandemic by the WHO on March 11th, 2020. ” In: Website of the RKI. Robert Koch Institute (RKI), May 26, 2020, accessed on May 26, 2020 .
- ↑ Information on the detection, diagnosis and therapy of COVID-19 patients (PDF). Ed .: " Standing Working Group of Competence and Treatment Centers for Diseases caused by Highly Pathogenic Pathogens " (STAKOB) - Office at the Robert Koch Institute , as of April 17, 2020, accessed on April 30, 2020.
- ↑ Neeltjevan Doremalen, Dylan H.Morris, Myndi G.Holbrook et al .: Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1 The New England Journal of Medicine, May 2020th
- ↑ Virologist Drosten on aerosol transmission - "In everyday life, concentrate more on airing than on constant disinfection". Retrieved May 25, 2020 .
- ↑ New Findings on Superspreader Events - A Summary of Current Studies by Klaus Taschwer. Retrieved May 26, 2020 .
- ↑ Covid-19: What role do “super spreaders” play in the spread of the coronavirus? Deutschlandfunk - Online , May 28, 2020, accessed on May 28, 2020 .
- ↑ Stephanie Hegarty: The Chinese doctor who tried to warn others about coronavirus. BBC News, February 6, 2020, accessed February 6, 2020 .
- ^ Deutsche Welle (www.dw.com): Whistleblower doctor from Wuhan dies | DW | 02/07/2020. Retrieved May 12, 2020 .
- ↑ a b c d Na Zhu, Dingyu Zhang, Wenling Wang, Xingwang Li, Bo Yang, Jingdong Song, Xiang Zhao, Baoying Huang, Weifeng Shi, Roujian Lu, Peihua Niu, Faxian Zhan, Xuejun Ma, Dayan Wang, Wenbo Xu, Guizhen Wu, George F. Gao, Wenjie Tan for the China Novel Coronavirus Investigating and Research Team: A Novel Coronavirus from Patients with Pneumonia in China, 2019 . In: The New England Journal of Medicine . January 24, 2020, doi : 10.1056 / NEJMoa2001017 (English).
- ^ Pneumonia of unknown cause - China. In: WHO website. January 5, 2020, accessed on January 14, 2020 .
- ↑ New virus surging in Asia rattles scientists. In: nature .com. January 20, 2020, accessed on January 28, 2020 .
- ↑ Wuhan seafood market may not be source of novel virus spreading globally. In: ScienceMag of the AAAS. January 26, 2020, accessed on April 23, 2020 .
- ↑ Chaolin Huang, Yeming Wang, Xingwang Li, Lili Ren, Jianping Zhao, Yi Hu et al .: Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China , in: The Lancet Volume 395, No. 10223 of February 15 2020, pp. 497-506, published January 24, 2020, doi: 10.1016 / S0140-6736 (20) 30183-5
- ^ WHO Statement Regarding Cluster of Pneumonia Cases in Wuhan, China. WHO, January 9, 2020, accessed January 14, 2020 .
- ↑ a b Answers to frequently asked questions about the SARS-CoV-2 coronavirus. In: Website of the Robert Koch Institute . March 18, 2020, accessed March 19, 2020 .
- ↑ a b c F. Wu, S. Zhao, B. Yu, Y.-M. Chen, W. Wang, Y. Hu, Z.-G. Song, Z.-W. Tao, J.-H. Tian, Y.-Y. Pei, ML Yuan, Y.-L. Zhang, F.-H. Dai, Y. Liu, Q.-M. Wang, J.-J. Zheng, L. Xu, EC Holmes, Y.-Z. Zhang: Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1, complete genome. In: Nucleotide website of the National Center for Biotechnology Information (NCBI). Retrieved February 4, 2020 .
- ↑ WHO: Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. On: who.int from January 14, 2020.
- ↑ Karola Neubert: The first test for the novel corona virus in China has been developed. In: Website Informationsdienst Wissenschaft (idw). January 16, 2020, accessed February 9, 2020 .
- ↑ a b c d Victor M. Corman, Tobias Bleicker, Sebastian Brünink, Christian Drosten, Olfert Landt, Marion PG Koopmans, Maria Zambon, Malik Peiris: Diagnostic detection of 2019-nCoV by real-time RT-PCR (protocol-v2- 1) . Ed .: Charité Virologie, Berlin. January 17, 2019, p. 1–13 ( PDF, 1, MB [accessed February 1, 2020]).
- ↑ a b Unlocking the Genetic Code of the Novel Coronavirus: How COVID-19 Made the Leap From Animals to Humans , on SciTechDaily of March 26, 2020, source: University of Sydney
- ↑ a b c d e f g h i j k l Alexandre Hassanin: Coronavirus Could Be a 'Chimera' of Two Different Viruses, Genome Analysis Suggests , on: science alert of March 24, 2020 (Source: The Conversation)
- ↑ Aylin Woodward: Chinese CDC Now Says The Wuhan Wet Market Wasn't The Origin of The Virus , on: science alert of May 29, 2020, source: Business Insider
- ↑ Aylin Woodward: The Chinese CDC now says the coronavirus didn't jump to people at the Wuhan wet market - instead, it was the site of a superspreader event , on Business Insider (BI) of May 29, 2020
- ↑ https://www.sciencedirect.com/science/article/pii/S0924857920301643
- ↑ press release. (PDF) Albert Schweitzer Clinic Colmar, May 7, 2020, accessed on June 7, 2020 .
- ↑ Text of the regulation on the extension of the reporting obligation according to § 6 paragraph 1 sentence 1 number 1 and § 7 paragraph 1 sentence 1 of the Infection Protection Act to infections with the novel coronavirus that first appeared in Wuhan / People's Republic of China in December 2019 ("2019-nCoV")
- ↑ New coronavirus. In: bag.admin.ch. Federal Office of Public Health FOPH, March 13, 2020, accessed on March 15, 2020 (Swiss Standard German).
- ↑ Almost 100 more deaths from Covid-19 in China. Süddeutsche Zeitung , February 12, 2020, accessed on February 13, 2020 (direct from the dpa news channel).
- ↑ Novel Coronavirus (2019-nCoV). ( Memento from January 28, 2020 in the Internet Archive )
- ↑ Taxonomy ID: 2697049 Wuhan seafood market pneumonia virus. ( Memento from February 3, 2020 in the Internet Archive )
- ↑ Ching-Tse Cheng: WHO declines to name new pneumonia after 'China' or 'Wuhan'. Taiwan News, January 14, 2020, accessed January 14, 2020 .
- ^ WHO issues best practices for naming new human infectious diseases. World Health Organization, May 8, 2015, accessed February 6, 2020 .
- ↑ Taxonomy Browser: Severe acute respiratory syndrome coronavirus 2, Taxonomy ID: 2697049. In: Website National Center for Biotechnology Information (NCBI). Retrieved February 16, 2020 .
- ↑ Nicky Phillips, Smriti Mallapaty, David Cyranoski: How quickly does the Wuhan virus spread? In: Nature . January 21, 2020, doi : 10.1038 / d41586-020-00146-w (English).
- ↑ a b Lars Fischer, Alina Schadwinkel: Does the coronavirus cause drug shortages? Does the virus come from the pangolin? Website Spektrum.de , February 10, 2020, accessed on February 15, 2020 .
- ↑ Shibo Jiang, Zhengli Shi, Yuelong Shu, Jingdong Song, George F. Gao, Wenjie Tan, Deyin Guo: A distinct name is needed for the new coronavirus . In: The Lancet . February 19, 2020, doi : 10.1016 / S0140-6736 (20) 30419-0 (English).
- ↑ a b c d Kristian G. Andersen, Andrew Rambaut, W. Ian Lipkin, Edward C. Holmes, Robert F. Garry: The Proximal Origin of SARS-CoV-2. In: virological.org , Source: ARTIC Network, February 17, 2020, Nature
- ↑ a b Ben Hu, Lei-Ping Zeng, Xing-Lou Yang, Xing-Yi Ge, Wei Zhang et al .: Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. In: PLOS Pathogens , November 30, 2017, doi: 10.1371 / journal.ppat.1006698
- ↑ ICTV Master Species List 2018b.v2 . MSL # 34, March 2019
- ↑ a b c Xingguang Li, Junjie Zai, Qiang Zhao, Qing Nie, Yi Li, Brian T. Foley, Antoine Chaillon: Evolutionary history, potential intermediate animal host, and cross ‐ species analyzes of SARS ‐ CoV ‐ 2 , in: Journal of Medical Virology , February 27, 2020, doi: 10.1002 / jmv.25731 , PDF , PMID 32104911 , reseachGate
- ↑ Xiaolu Tang, Changcheng Wu, Xiang Li, Yuhe Song, Xinmin Yao, Xinkai Wu, Yuange Duan, Hong Zhang, Yirong Wang, Zhaohui Qian, Jie Cui, Jian Lu: On the origin and continuing evolution of SARS-CoV-2 , on: National Science Review (NSR, Oxford Academic) of March 3, 2020, nwaa036, doi: 10.1093 / nsr / nwaa036
- ↑ Scientists discover two main subtypes of the novel coronavirus , on: People's Dayly Online, China (German)
- ↑ Jessica Hamzelou: Coronavirus: Are there two strains and is one more deadly? , on NewScientist of March 5, 2020 (with coronavirus here only SARS-CoV-2 is meant - free article)
- ↑ SARS-CoV-2: Are there 2 different virus strains? , on: aerzteblatt.de of March 9, 2020
- ↑ Are there two strains of Sars-CoV-2? , on: n-tv.de from March 10, 2020
- ↑ Korinna Hennig, Christian Drosten: Coronavirus update: "Viruses always mutate" , interview on ndr.de from March 6, 2020
- ↑ Peter Forster, Lucy Forster, Colin Renfrew, Michael Forster: Phylogenetic network analysis of SARS-CoV-2 genomes , on: PNAS of April 8, 2020, doi: 10.1073 / pnas.2004999117
- ↑ Genetic Study Identifies Three Variants of SARS-CoV-2 Coronavirus , on: Sci-News of April 9, 2020
- ↑ Kai Kupferschmidt: The family tree of the pandemic , on: Spektrum.de from March 18, 2020 (with reference to Christian Drosten and Andrew Rambaut)
- ↑ Amanda Woods: Iceland scientists found 40 mutations of the coronavirus, report says , on: New Yor Post of March 24, 2020
- ↑ Bo Elkjær: Forskere har sporet 40 mutationer af coronavirus - alene på Iceland , on: www.information.dk (Iceland outlet Information) of March 24, 2020, Danish
- ↑ Vanessa Chalmers: Scientists in Iceland claim they have found FORTY mutations of the coronavirus - and admit seven cases can be traced back to 'a football match in England' , on: www.dailymail.co.uk (MailOnline) from March 24th 2020
- ↑ Alexander Elliott: Two types of COVID-19 in one individual , on: www.ruv.is (RÚV news), Iceland , from March 24, 2020
- ↑ Poppy Askham: Patient Infected With Two Strains of COVID-19 In Iceland , on: The Reykjavík Grapevine, March 24, 2020
- ↑ a b c Lucy van Dorp, Mislav Acmana, Damien Richard, Liam P. Shawd, Charlotte E. Ford, Louise Ormond, Christopher J. Owen, Juanita Pang, Cedric C. S. Tan, Florencia A. T. Boshier, Arturo Torres Ortiz, François Ballou: Emergence of genomic diversity and recurrent mutations in SARS-CoV-2 , in: In Press, Journal Pre-proof, doi: 10.1016 / j.meegid.2020.104351
- ↑ SARS-CoV-2: Mutations could further increase infectiousness , aerzteblatt.de of May 6, 2020
- ↑ Nadja Podbregar: How strongly has the coronavirus mutated? , on scinexx.de on May 8, 2020
- ^ Bette Korber, David D. Montefiori et. al. : Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2 , bioarxiv April 30, 2020
- ↑ Confirmed: New variant "infectious" , on: science.orf.at of July 3, 2020
- ↑ Nadja Podbregar: A mutated form of SARS-CoV-2 has almost replaced the original variant , on: scinexx.de of July 3, 2020: D614 versus G614
- ↑ Bette Korber, W. M. Fischer, S. Gnanakaran, E. O. Saphire, D. C. Montefiori et al. : Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus , in: Cell from July 2, 2020, doi: 10.1016 / j.cell.2020.06.043
- ↑ More "spines" through mutation - is the coronavirus becoming more contagious? , on: n-tv.de from June 15, 2020
- ↑ a b c d e NCBI Database Nucleotide, txid2697049 (Severe acute respiratory syndrome coronavirus 2). In: Nucleotide website of the National Center for Biotechnology Information (NCBI). Accessed February 16, 2020 .
- ^ Matthew Frieman, Ralph Baric: Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation . In: Microbiology and Molecular Microbiology Reviews . tape 72 , December 2008, p. 672-685 , doi : 10.1128 / MMBR.00015-08 , PMID 19052324 (English, open access).
- ^ Wuhan seafood market pneumonia virus. In: National Center for Biotechnology Information (NCBI) Genome website . Accessed February 16, 2020 .
- ↑ a b c d e Jasper Fuk-Woo Chan, Shuofeng Yuan, Kin-Hang Kok, Kelvin Kai-Wang To, Hin Chu, Jin Yang, Fanfan Xing, Jieling Liu, Cyril Chik-Yan Yip, Rosana Wing-Shan Poon, Hoi-Wah Tsoi, Simon Kam-Fai Lo, Kwok-Hung Chan, Vincent Kwok-Man Poon, Wan-Mui Chan, Jonathan Daniel Ip, Jian-Piao Cai, Vincent Chi-Chung Cheng, Honglin Chen, Christopher Kim-Ming Hui , Kwok-Yung Yuen: A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster . In: The Lancet . January 24, 2020, doi : 10.1016 / S0140-6736 (20) 30154-9 (English).
- ↑ a b Horseshoe bat bats. Protection Association of German Forests , Oberursel from December 16, 2015
- ↑ a b c Peng Zhou, Xing-Lou Yang, Xian-Guang Wang, Ben Hu, Lei Zhang, Wei Zhang, Hao-Rui Si, Yan Zhu, Bei Li, Chao-Lin Huang, Hui-Dong Chen, Jing Chen, Yun Luo, Hua Guo, Ren-Di Jiang, Mei-Qin Liu, Ying Chen, Xu-Rui Shen, Xi Wang, Xiao-Shuang Zheng, Kai Zhao, Quan-Jiao Chen, Fei Deng, Lin-Lin Liu, Bing Yan , Fa-Xian Zhan, Yan-Yi Wang, Geng-Fu Xiao, Zheng-Li Shi: A pneumonia outbreak associated with a new coronavirus of probable bat origin . In: Nature . February 3, 2020, doi : 10.1038 / s41586-020-2012-7 (English, this article was published on bioRxiv on January 23, 2020 in advance without peer review).
- ↑ D. Paraskevis, EG Kostaki, G. Magiorkinis, G. Panayiotakopoulos, G. Sourvinos, S. Tsiodras: Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event . In: bioRxiv . January 27, 2020, doi : 10.1101 / 2020.01.26.920249 (English).
- ↑ a b c d Roujian Lu, Xiang Zhao, Juan Li, Peihua Niu, Bo Yang, Honglong Wu, Wenling Wang, Hao Song, Baoying Huang, Na Zhu, Yuhai Bi, Xuejun Ma, Faxian Zhan, Liang Wang, Tao Hu, Hong Zhou, Zhenhong Hu, Weimin Zhou, Li Zhao, Jing Chen, Yao Meng, Ji Wang, Yang Lin, Jianying Yuan, Zhihao Xie, Jinmin Ma, William J Liu, Dayan Wang, Wenbo Xu, Edward C Holmes, George F Gao , Guizhen Wu, Weijun Chen, Weifeng Shi, Wenjie Tan: Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding . In: The Lancet . January 29, 2020, doi : 10.1016 / S0140-6736 (20) 30251-8 (English).
- ↑ Genome analyzes clarify the origin of 2019-nCoV. In: Website Deutsches Ärzteblatt . January 30, 2020, accessed February 9, 2020 .
- ↑ 2019-nCoV: First pictures of the virus and findings on the clinical course. In: Website Deutsches Ärzteblatt . January 27, 2020, accessed February 11, 2020 .
- ↑ https://link.springer.com/content/pdf/10.1007%2F978-3-8274-2241-5_14.pdf
- ^ Qiu, Y .; Zhao, Y .; Wang, Q .; Li, J .; ZHou, Z .; Liao, C .; Ge, X. Predicting the Angiotensin Converting Enzyme 2 (ACE2) Utilizing Capability as the Receptor of SARS-CoV-2. Preprints 2020, 2020030091 (doi: 10.20944 / preprints202003.0091.v1)
- ↑ Christian JA Sigrist, Alan Bridge, Philippe Le Mercier: A potential role for integrins in host cell entry by SARS-CoV-2 . In: Antiviral Research . tape 177 , May 2020, p. 104759 , doi : 10.1016 / j.antiviral.2020.104759 ( elsevier.com [accessed April 30, 2020]).
- ↑ Hoffmann et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a ClinicallyProven Protease Inhibitor, Cell (2020), https://doi.org/10.1016/j.cell.2020.02.052
- ↑ Blake Oberfeld, Aditya Achanta, Kendall Carpenter, Pamela Chen, Nicole M. Gilette: SnapShot: COVID-19 . In: Cell . tape 181 , no. 4 , May 2020, p. 954–954.e1 , doi : 10.1016 / j.cell.2020.04.013 , PMC 7190493 (free full text) - ( elsevier.com [accessed May 15, 2020]).
- ↑ Adedeji, Adeyemi & Severson, William & Jonsson, Colleen & Singh, Kamalendra & Weiss, Susan & Sarafianos, Stefan. (2013). Novel Inhibitors of SARS-CoV Entry acting by Three Distinct Mechanisms. Journal of virology. 87.10.1128 / JVI.00998-13.
- ↑ RNA viruses. Spektrum.de, accessed on August 9, 2020 .
- ↑ http://hdl.handle.net/2268/4138
- ↑ Erwan Sallard, Jose Halloy, Didier Casane, Etienne Decroly, Jacques vanHelden: Tracing the origins of SARS-COV-2 in coronavirusphylogenies , on: HAL archives-ouvertes, Version 2 (CCSD), July 6, 2020, HAL Id: hal -02891455
-
↑ Joana Damas, Graham M. Hughes, Kathleen C. Keough, Corrie A. Painter, Nicole S. Persky, Marco Corbo, Michael Hiller, Klaus-Peter Koepfli, Andreas R. Pfenning, Huabin Zhao, Diane P. Genereux, Ross Swofford , Katherine S. Pollard, Oliver A. Ryder, Martin T. Nweeia, Kerstin Lindblad-Toh, Emma C. Teeling, Elinor K. Karlsson, Harris A. Lewin; Scott V. Edwards (Ed.): Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates , in: PNAS from August 21, 2020, doi: 10.1073 / pnas.2010146117 .
Many Animal Species Vulnerable to SARS-CoV-2 / COVID-19 According to Genomic Analysis , on: SciTechDaily, August 23, 2020, Source: University of California - Davis.
Nadja Podbregar: Corona: Which animal species are susceptible? , on: scinexx.de from August 24, 2020. - ↑ a b Ewen Callaway, David Cyranoski: Why snakes probably aren't spreading the new China virus - One genetic analysis suggests reptilian reservoir - but researchers doubt that the coronavirus could have originated in animals other than birds or mammals . In: Nature . January 23, 2020, doi : 10.1038 / d41586-020-00180-8 (English).
- ↑ Wei Ji, Wei Wang, Xiaofang Zhao, Junjie Zai, Xingguang Li: Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross ‐ species transmission from snake to human . In: Journal of Medical Virology . January 22, 2020, doi : 10.1002 / jmv.25682 (English).
- ↑ Researchers trace coronavirus outbreak in China to snakes. In: Website EurekAlert! January 22, 2020, accessed on January 26, 2020 .
- ↑ Coronavirus: How worried should we be? BBC News , January 27, 2020, accessed January 27, 2020 .
- ↑ a b c Chengxin Zhang et al .: Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1 , in: American Chemical Society: J. Proteome Res . of March 22, 2020, doi: 10.1021 / acs.jproteome.0c00129 ; PrePrint , PrePrint full text (PDF) from February 8, 2020
- ↑ NCBI: Bat coronavirus RaTG13 (no rank)
- ↑ a b Jose Halloy, Erwan Sallard, José Halloy, Didier Casane, Etienne Decroly, Jacques van Helden: Tracing the origins of SARS-COV-2 in coronavirus phylogenies , in: HAL from July 16, 2020, HAL Id: hal-02891455 (Preprint)
- ↑ David Cyranoski: Did pangolins spread the China coronavirus to people? In: Nature . February 7, 2020, doi : 10.1038 / d41586-020-00364-2 (English).
- ↑ Mike McRae: Coronaviruses Similar to The COVID-19 One Have Just Been Found in Pangolins , on science alert of March 27, 2020 (with "COVID-19" is not the human disease, but it generally means Sarbecoviruses, with "Coronaviruses “Specifically only SARS-CoV-2). The pangolins or parts of them had been confiscated by the Chinese customs, at Pan_SL-CoV_GD in the Guandong Province , at Pan_SL-CoV_GX in the Guangxi Province .
- ↑ Tommy Tsan-Yuk Lam et al .: Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins , in: Nature from March 26, 2020, doi: 10.1038 / s41586-020-2169-0 (preprint)
- ↑ Pangolins, Not Snakes, May Be Missing Link in Coronavirus Jump From Bats to Humans , on: SciTechDaily of March 27, 2020, source: American Chemical Society
- ↑ Tina Hesman Saey: https://www.sciencenews.org/article/coronavirus-covid-19-not-human-made-lab-genetic-analysis-nature , on ScienceNews of March 26, 2020
- ↑ Xiaojun Li, Elena E. Giorgi, Manukumar Honnayakanahalli Marichannegowda, Brian Foley, Chuan Xiao, Xiang-Peng Kong, Yue Chen, S. Gnanakaran, Bette Korber, Feng Gao: Emergence of SARS-CoV-2 through recombination and strong purifying selection , in: ScienceAdvances, AAAS, from May 29, 2020, eabb9153, doi: 10.1126 / sciadv.abb9153
- ↑ Bats, Pangolins and Humans: COVID-19 Virus Likely Emerged From Recombination of Viral Genes Across Different Species , on: ScitechDaily from May 31, 2020. Quote: “ … the virus' entire receptor binding motif (RBM), a component that plays a key role in viral entry into host cells, which was introduced [into specific bat coronaviruses] through recombination with pangolin coronaviruses. "
- ↑ Evolution of Pandemic Coronavirus Outlines Path From Animals to Humans - Highlights Future Danger , on: SciTechDaily from June 6, 2020, source: DUKE UNIVERSITY MEDICAL CENTER
- ↑ Rachel L. Graham, Ralph S. Baric: Recombination, Reservoirs, and the Modular Spike: Mechanisms of Coronavirus Cross-Species Transmission , in: ASM: Journal of Virology 84 (7), March 2010, pp. 3134-3146, doi : 10.1128 / JVI.01394-09 , PDF
- ↑ David Cyranoski: Virology: Portrait of a Killer , online edition of the article in Spectrum of Science No. 8, August 2020, pp. 40–49
-
↑ Maciej F. Boni, Philippe Lemey, Xiaowei Jiang, Tommy Tsan-Yuk Lam, Blair W. Perry, Todd A. Castoe, Andrew Rambaut, David L. Robertson: Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic , in: Nature Microbiology of July 28, 2020, doi: 10.1038 / s41564-020-0771-4 ; on:
Nadja Podbregar: On the trail of Sars-CoV-2 , on: Wissenschaft.de from July 28, 2020
Nadja Podbregar: Corona: SARS-CoV-2 has been around for decades , on: scinexx.de from 29 July 2020
Erin Garcia de Jesus: Close relatives of the coronavirus may have been in bats for decades , on: ScienceNews of July 28, 2020 - ↑ Raccoon dogs as intermediate hosts? Drosten brings a new virus source into play , on n-tv.de from April 26, 2020
- ↑ The raccoon dog as a coronavirus sling?
- ↑ James D. Cherry, Paul Krogstad: SARS: The First Pandemic of the 21st Century
- ↑ Q&A on coronaviruses (COVID-19). Retrieved March 6, 2020 .
- ↑ a b Coronavirus: No, dogs do not get Covid-19. In: Der Spiegel - Wissenschaft. Retrieved March 6, 2020 .
- ↑ Coronaviruses can be dangerous for pets. In: Frankfurter Neue Presse. March 6, 2020, accessed March 6, 2020 .
- ↑ Immediate notification. Tai Hang, Islands District, Hong Kong. On: oie.int , World Organization for Animal Health (OIE), February 29, 2020.
- ↑ Noah Higgins-Dunn: A dog in Hong Kong tests positive for the coronavirus, WHO officials confirm. In: CNBC. February 28, 2020, accessed on February 29, 2020 .
- ↑ World Organization for Animal Health (OIE): Immediate notification. Tai Hang, Islands District, Hong Kong. On: oie.int on February 29, 2020.
- ↑ Follow-up report No. 3 (Final report): COVID-19 (SARS-COV-2), Hong Kong. Likely human to animal transmission. On: oie.int on March 28, 2020.
- ↑ Immediate notification. Pok Fu Lam, Southern District, Hong Kong. On: oie.int , World Organization for Animal Health (OIE), March 21, 2020.
- ↑ Xuhua Xia: Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense , in: Molecular Biologa and Evolution, Academic Press, April 14, 2020, doi: 10.1093 / molbev / msaa094
- ↑ Evidence of Stray Dogs as Possible Origin of COVID-19 Pandemic , on: SciTechDaily from April 14, 2020, source: University of Ottawa
- ↑ Case in Belgium - cat tested positive for coronavirus , on n-tv.de from March 27, 2020.
- ^ Coronavirus: Belgian cat infected by owner. On: brusselstimes.com of March 27, 2010.
- ^ Report of the Agriculture, Fisheries and Conservation Department, Hong Kong. On: oie.int , World Organization for Animal Health (OIE), May 5, 2020.
- ^ Qiang Zhang et al .: SARS-CoV-2 neutralizing serum antibodies in cats: a serological investigation. On: bioRxiv , preprint of April 3, 2020, doi: 10.1101 / 2020.04.01.021196 .
- ↑ coronavirus can infect cats - dogs not so much. On: nature.com from April 1, 2020.
- ↑ Peter J. Halfmann et al. : Transmission of SARS-CoV-2 in Domestic Cats. In: The New England Journal of Medicine , online publication May 13, 2020, doi: 10.1056 / NEJMc2013400 .
- ↑ Cats Can Spread COVID-19 Coronavirus Infection to Other Cats , on: SciTechDaily of March 29, 2020, source: University of Tokyo
- ↑ a b Lars Fischer: Coronavirus: What role do pets play in the pandemic? , on: Spektrum.de from June 4, 2020, Source: Nature
- ^ A Tiger In New York Has tested Positive For Coronavirus. On: sciencealert.com April 6, 2020. Source: Agence France-Presse.
- ↑ OIE: Immediate notification: SARS-CoV-2 / COVID-19, United States of America. Bronx County, Bronx, New York. On: oie.int from April 6, 2020.
- ↑ SARS-COV-2 / COVID-19, South Africa. Report from the Department of Agriculture, Forestry and Fisheries, Animal Production and Health to the OIE, August 11, 2020.
- ↑ Young-Il Kim et al .: Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. In: Cell Host & Microbe , online pre-publication of April 6, 2020, doi: 10.1016 / j.chom.2020.03.023 .
- ↑ New coronavirus SARS-CoV-2: fruit bats and ferrets are susceptible, pigs and chickens are not. On: idw-online.de from April 2, 2020.
- ↑ Jianzhong Shi et al. : Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS – coronavirus 2. In: Science , online advance publication of April 8, 2020, eabb7015, doi: 10.1126 / science.abb7015 .
- ^ Jon Cohen: From mice to monkeys, animals studied for coronavirus answers. In: Science , Volume 368, No. 6488, 2020, pp. 221 f, doi: 10.1126 / science.368.6488.221 , full text
- ↑ Coronavirus rips through Dutch mink farms, triggering culls to prevent human infections. On: sciencemag.org June 9, 2020.
- ↑ Fur animals fall ill with Covid-19 en masse. On: sueddeutsche.de from May 14, 2020.
- ↑ Nadia Oreshkova et al .: SARS-CoV2 infection in farmed mink, Netherlands, April 2020. Preprint on bioRxiv from May 18, 2020, doi: 10.1101 / 2020.05.18.101493 .
- ↑ International Society for Infectious Diseases: Coronavirus Disease 2019 Update (209): Netherlands (North Brabant), Farmed Mink, Animal-to-Human, Cat, Epidemiology. Report of May 25, 2020.
- ↑ International Society for Infectious Diseases: Coronavirus Disease 2019 Update (215): Netherlands (NB), Animal, Mink-to-Human, Epidemiology, Control. Report of May 27, 2020.
- ↑ Kore Schlottau, Melanie Rissmann, Annika Graaf, Jacob Schön, Julia Sehl, Claudia Wylezich, Martin Beer et al. : SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study , in: The Lancet from 7 Jul 2020, doi: https: //doi.org/10.1016/S2666-5247 (20) 30089-6 , PDF
- ↑ Shi-Hui Sun, You-Chun Wang et al. : A Mouse Model of SARS-CoV-2 Infection and Pathogenesis , in: Cdell Hust and Microbe from May 26, 2020, doi: 10.1016 / j.chom.2020.05.020
- ↑ Jacinta Bowler: Scientists Find a Way to Infect Mice With Coronavirus. Here's Why That's So Important , on: science alert from June 1, 2020
- ↑ Andrea J. Pruijssers et al. : Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice , in: Cell Reports from July 7, 2020, doi: 10.1016 / j.celrep.2020.107940 (free Pre-proof)
- ↑ Tina Hesman Saey: Remdesivir may work even better against COVID-19 than we thought , on: ScienceNews of July 13, 2020
- ↑ COVID-19: protect great apes during human pandemics. In: Nature , Volume 579, 2020, p. 497, doi: 10.1038 / d41586-020-00859-y .
- ↑ NCBI: Human coronavirus OC43 (no rank)
- ↑ a b Nadja Podbregar: Coronavirus: Are great apes also endangered? On: scinexx.de from March 30, 2020.
- ↑ Elle Hunt: Orangutans and other great apes under threat from covid-19 pandemic , on: NewScientist, April 2, 2020
- ↑ Chuan Qin et al .: Reinfection could not occur in SARS-CoV-2 infected rhesus macaques . In: bioRxiv . March 14, 2020, bioRxiv : 10.1101 / 2020.03.13.990226v1 ( preprint - full text), doi : 10.1101 / 2020.03.13.990226 .
- ↑ No multiple SARS-CoV-2 infection in monkeys. (No longer available online.) In: Deutsches Ärzteblatt . Deutscher Ärzteverlag , March 18, 2020, archived from the original on March 19, 2020 ; accessed on March 19, 2020 .
- ↑ Can people get Covid-19 twice? , on: n-tv.de of April 18, 2019, source: ntv.de, Amélie Bottollier-Depois, AFP
- ↑ Abishek Chandrashekar et al .: SARS-CoV-2 infection protects against rechallenge in rhesus macaques. In: Science , online advance publication May 20, 2020, eabc4776, doi: 10.1126 / science.abc4776 .
- ↑ Barry Rockx, Thijs Kuiken, Sander Herfst et al .: Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. In: Science , online advance publication of April 17, 2020, abb7314, doi: 10.1126 / science.abb7314 .
- ↑ Information from the BAuA: Novel virus SARS-CoV-2 (previously 2019-nCoV) classified by the ABAS in risk group 3 and recommendations for laboratory diagnostics are given. In: Website of the Federal Institute for Occupational Safety and Health (BAuA). February 19, 2020, accessed February 23, 2020 .
- ↑ a b Decision 1/2020 of the ABAS of February 19, 2020 and justification for the preliminary classification of the SARS-CoV-2 virus in risk group 3 and recommendations for non-targeted activities (laboratory diagnostics) and targeted activities with SARS-CoV-2. (PDF; 140 kB) In: Website of the Federal Institute for Occupational Safety and Health . February 19, 2020, accessed February 23, 2020 .
- ↑ Interim Laboratory Biosafety Guidelines for Handling and Processing Specimens Associated with Coronavirus Disease 2019 (COVID-19). In: Website of the US Centers for Disease Control and Prevention (CDC). February 16, 2020, accessed on February 23, 2020 .
- ↑ a b c Victor M. Corman, Tobias Bleicker, Sebastian Brünink, Christian Drosten, Olfert Landt, Marion PG Koopmans, Maria Zambon, Malik Peiris: Diagnostic detection of 2019-nCoV by real-time RT-PCR (protocol-v2-1 ) . Ed .: Charité Virologie, Berlin. January 17, 2019 ( PDF, 1, MB [accessed April 10, 2020]).
- ↑ a b c d e Victor M. Corman, Olfert Landt, Marco Kaiser, Richard Molenkamp, Adam Meijer, Daniel KW Chu, Tobias Bleicker, Sebastian Brünink, Julia Schneider, Marie Luisa Schmidt, Daphne GJC Mulders, Bart L. Haagmans, Bas van der Veer, Sharon van den Brink, Lisa Wijsman, Gabriel Goderski, Jean-Louis Romette, Joanna Ellis, Maria Zambon, Malik Peiris, Herman Goossens, Chantal Reusken, Marion PG Koopmans, Christian Drosten: Detection of 2019 novel coronavirus (2019- nCoV) by real-time RT-PCR . In: Eurosurveillance . tape 25 , no. 3 , January 23, 2020, ISSN 1560-7917 , p. 1–8 , doi : 10.2807 / 1560-7917.ES.2020.25.3.2000045 (English).
- ↑ Paul Ehrlich Institute: COVID-19 tests: NAT test is considered the gold standard , NAT tests also include the PCR method. As of March 23, 2020, accessed on August 25, 2020.
- ↑ a b c d e Coronavirus outbreak: New test kit can detect the virus in just 15 minutes. In: MSN . February 10, 2020, accessed on February 16, 2020 .
- ↑ a b Victor M. Corman, Tobias Bleicker, Sebastian Brünink, Christian Drosten, Olfert Landt, Marion PG Koopmans, Maria Zambon, Malik Peiris: Diagnostic detection of 2019-nCoV by real-time RT-PCR (protocol-v2-1) . Ed .: Charité Virologie, Berlin. January 17, 2019, p. 1 below (English, PDF, 1, MB [accessed April 10, 2020]).
- ^ Producer of corona tests: company in a state of emergency. In: The daily newspaper taz.de. March 12, 2020, accessed March 14, 2020 .
- ↑ Coronavirus disease (COVID-19) technical guidance: Laboratory testing for 2019-nCoV in humans. In: WHO website. World Health Organization (WHO), 2020, accessed March 15, 2020 .
- ↑ Stefan Nicola: A Berlin Biotech Company Got a Head Start on Coronavirus Tests. In: Bloomberg Businessweek . March 12, 2020, accessed on March 14, 2020 .
- ↑ Frankfurt researchers develop faster coronavirus test. In: bz-berlin.de. March 31, 2020, accessed April 1, 2020 .
- ↑ Direct pathogen detection using RT-PCR. In: Website of the Robert Koch Institute . August 11, 2020, accessed August 30, 2020 .
- ↑ Victor M. Corman, Tobias Bleicker, Sebastian Brünink, Christian Drosten, Olfert Landt, Marion PG Koopmans, Maria Zambon, Malik Peiris: Diagnostic detection of 2019-nCoV by real-time RT-PCR (protocol-v2-1) . Ed .: Charité Virologie, Berlin. January 17, 2019, p. 3 , in the description of "Figure 2" (English, who.int [PDF; 1000 kB ; accessed on April 23, 2020]).
- ↑ Victor M. Corman, Tobias Bleicker, Sebastian Brünink, Christian Drosten, Olfert Landt, Marion PG Koopmans, Maria Zambon, Malik Peiris: Diagnostic detection of 2019-nCoV by real-time RT-PCR (protocol-v2-1) . Ed .: Charité Virologie, Berlin. January 17, 2019, p. 6 , title 1, paragraph 3 (English, who.int [PDF; 1000 kB ; accessed on April 23, 2020]).
- ↑ Patrick DM, Petric M, Skowronski DM, Guasparini R, Booth TF, Krajden M, McGeer P, Bastien N, Gustafson L, Dubord J, Macdonald D, David ST, Srour LF, Parker R, Andonov A, Isaac-Renton J. , Loewen N, McNabb G, McNabb A, Goh SH, Henwick S, Astell C, Guo JP, Drebot M, Tellier R, Plummer F, Brunham RC: An Outbreak of Human Coronavirus OC43 Infection and Serological Cross-reactivity with SARS Coronavirus . In: The Canadian journal of infectious diseases & medical microbiology = Journal canadien des maladies infectieuses et de la microbiologie medicale . tape 17 , no. 6 , November 2006, pp. 330–336 , doi : 10.1155 / 2006/152612 , PMID 18382647 , PMC 2095096 (free full text) - (English).
- ↑ Prof. Dr. Heinz Konstruhardt, Dr. Martin Kammel: Comment on the extra interlaboratory comparison group 340 virus genome detection - SARS-CoV-2. (PDF) June 3, 2020, accessed June 10, 2020 .
- ↑ a b Foundation for Innovative New Diagnostics: SARS-CoV-2 molecular assay evaluation: results. June 9, 2020, accessed June 13, 2020 .
- ↑ Kerstin Wernike, Markus Keller, Franz J. Conraths, Thomas C. Mettenleiter, Martin H. Groschup, Martin Beer: Pitfalls in SARS-CoV-2 PCR Diagnostics . In: Transboundary and Emerging Diseases . June 14, 2020, doi : 10.1111 / tbed.13684 (English).
- ↑ a b Yafang Li, Lin Yao, Jiawei Li, Lei Chen, Yiyan Song, Zhifang Cai, Chunhua Yang: Stability issues of RT-PCR testing of SARS- CoV-2 for hospitalized patients clinically diagnosed with COVID-19 . In: Journal of medical virology . March 26, 2020, doi : 10.1002 / jmv.25786 , PMID 32219885 (English, Epub ahead of print).
- ↑ a b Emily Feng, Amy Cheng: Mystery In Wuhan: Recovered Coronavirus Patients Test Negative ... Then Positive. In: NPR.org . National Public Radio , Inc. (USA), March 27, 2020, accessed April 14, 2020 (English, partly misleading: The sentence: “ There are false positives with these types of tests, ” does not refer to a specific test, as one might guess).
- ↑ Jaffar Al-Tawfiq, Ziad A. Memish: Diagnosis of SARS- CoV-2 Infection based on CT scan vs. RT-PCR: Reflecting on Experience from MERS-CoV . In: The Journal of Hospital Infection . March 5, 2020, doi : 10.1016 / j.jhin.2020.03.001 (English).
- ↑ Interpreting a covid-19 test result , BMJ 2020; 369: m1808, online May 12, 2020, accessed August 30, 2020
- ↑ Cristina Helberg: Corona PCR test and pre-test probability: this can lead to incorrect results , online June 18, 2020, accessed August 28, 2020
- ↑ Spahn: much more false positives than actually positive , ARD, according to a report from Berlin, online June 14, 2020, access August 28, 2020
- ↑ Jens Spahn does not consider coronavirus tests to be effective for everyone , in: Die ZEIT, online June 19, 2020, accessed June 29, 2020
- ↑ ALM: Urgent appeal for resource-oriented use of the tests , online August 11, 2020, access August 22, 2020
- ↑ a b c d e Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. In: WHO website. World Health Organization (WHO), January 17, 2020, accessed February 5, 2020 .
- ↑ Global Initiative on Sharing All Influenza Data (GISAID). In: Website of GISAID (Federal Republic of Germany as host (database provider ) ). Retrieved February 5, 2020 .
- ↑ CDC Tests for 2019-nCoV. In: Website of the US Centers for Disease Control and Prevention (CDC). February 5, 2020, accessed February 10, 2020 .
- ↑ Stephanie Soucheray: CDC sends out nCoV test kits as Wisconsin confirms case. In: University of Minnesota website . CIDRAP - Center for Infectious Disease Research and Policy, February 5, 2020, accessed on February 10, 2020 .
- ↑ Axel Schröder: Corona crisis: This is how virus detection kits are produced. In: Website Deutschlandfunk . March 6, 2020, accessed March 15, 2020 .
- ↑ RealStar SARS-CoV-2 RT-PCR Kit 1.0. (PDF; 596 kB) In: Website Altona Diagnostics. February 2020, accessed on March 15, 2020 .
- ↑ a b c M. Parčina, UV Schneider, B. Visseaux, R. Jozić, I. Hannet, JG Lisby: Multicenter evaluation of the QIAstat Respiratory Panel - A new rapid, highly multiplexed PCR based assay for diagnosis of acute respiratory tract infections . In: PLOS ONE . tape 15 , no. 3 , March 12, 2020, p. e0230183 , doi : 10.1371 / journal.pone.0230183 , PMID 32163484 , PMC 7067435 (free full text).
- ↑ New test kits for coronavirus approved in China. In: Xinhua website . January 30, 2020, accessed February 10, 2020 .
- ↑ China's Jiangsu develops rapid test kit for coronavirus. In: Xinhua website . January 31, 2020, accessed on February 16, 2020 .
- ↑ Dean Koh: iHealthtech researchers working on Wuhan novel coronavirus (2019-nCoV) detection kit. In: National University of Singapore's mobi health news website . February 6, 2020, accessed on February 16, 2020 .
- ↑ M. Döhla, C. Boesecke, B. Schulte, C. Diegmann, E. Sib: Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity . In: Public Health . tape 182 , May 1, 2020, ISSN 0033-3506 , p. 170–172 , doi : 10.1016 / j.puhe.2020.04.009 , PMC 7165286 (free full text) - ( sciencedirect.com [accessed April 26, 2020]).
- ↑ a b c Hilden-based company develops rapid test for corona virus. In: wdr.de. February 5, 2020, accessed February 28, 2020 .
- ↑ a b c Press release: QIAGEN announces worldwide shipments of QIAstat-Dx test kits for SARS-CoV-2. In: Website Qiagen . February 26, 2020, accessed on February 28, 2020 .
- ↑ a b Maike Telgheder: Coronavirus diagnosis: The industry is developing rapid tests. In: Handelsblatt website . February 26, 2020, accessed February 28, 2020 .
- ↑ a b c d e f Andreas Stiller: A deeper insight into the infection tests against the SARS-CoV-2 coronavirus. In: heise online . March 27, 2020, accessed March 28, 2020 .
- ↑ a b c S. Y. Xiao, Y. Wu, H. Liu: Evolving status of the 2019 novel coronavirus infection: Proposal of conventional serologic assays for disease diagnosis and infection monitoring. In: Journal of Medical Virology. Volume 92, number 5, May 2020, published online in advance, pp. 464-467, doi: 10.1002 / jmv.25702 , PMID 32031264 .
- ↑ a b Juanjuan Zhao, Quan Yuan et al .: Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019 . In: Clinical Infectious Diseases . March 28, 2020, doi : 10.1093 / cid / ciaa344 .
- ↑ 2019-nCoV: Investigation of suspicions and measures - Orientation aid for doctors. In: Website of the Robert Koch Institute . January 23, 2020, accessed April 3, 2020 .
- ↑ Information on testing patients for infection with the novel coronavirus SARS-CoV-2. In: Website of the Robert Koch Institute (RKI). February 20, 2020, accessed February 25, 2020 .
- ↑ a b LADR informs Analysis No. 292 03/2020: Testing for antibodies against COVID-19 (SARS-CoV-2) available
- ↑ a b c d Nisreen MA Okba1, Marcel A. Müller, Wentao Li, Chunyan Wang, Corine H. GeurtsvanKessel, Victor M. Corman, Mart M. Lamers, Reina S. Sikkema, Erwin de Bruin, Felicity D. Chandler, Yazdan Yazdanpanah, Quentin Le Hingrat, Diane Descamps, Nadhira Houhou-Fidouh, Chantal BEM Reusken, Berend-Jan Bosch, Christian Drosten, Marion PG Koopmans, Bart L. Haagmans: Severe Acute Respiratory Syndrome Coronavirus 2 - Specific Antibody Responses in Coronavirus Disease 2019 Patients . In: Emerging Infectious Diseases . tape 26 , no. 7 , doi : 10.3201 / eid2607.200841 (July 2020, published online in advance).
- ↑ a b c d antibodies and their antigens; Methods of microbiological diagnosis . In: Helmut Hahn, Stefan HE Kaufmann, Thomas F. Schulz, Sebastian Suerbaum (eds.): Medical microbiology and infectious diseases . 6th edition. Springer Verlag, Heidelberg 2009, ISBN 978-3-540-46359-7 , p. 48-62, 140-150 .
- ↑ Some People May Have a Head Start Against Coronavirus, Surprising Evidence Shows , on: science alert from June 4, 2020, source: Business Insider
- ^ Jef Akst: Australian Lab Cultures New Coronavirus as Infections Climb. In: Website The-Scientist. January 29, 2020, accessed February 4, 2020 .
- ↑ Institut Pasteur isolates strains of coronavirus 2019-nCoV detected in France. In: Website EurekAlert! January 31, 2020, accessed February 4, 2020 .
- ↑ nCoV isolated in cell culture for the first time in Germany. In: Website Institute for Microbiology of the Bundeswehr . January 31, 2020, accessed February 3, 2020 .
- ↑ CDC Grows 2019-nCoV Virus in Cell Culture. In: Website of the US Centers for Disease Control and Prevention (CDC). February 6, 2020, accessed on February 10, 2020 .
- ^ Jon Cohen: Scientists are moving at record speed to create new coronavirus vaccines — but they may come too late . In: Science . January 27, 2020, doi : 10.1126 / science.abb0612 (English).
- ↑ Finn Mayer-Kuckuk: Search for corona vaccine: The hot race . In: The daily newspaper: taz . March 17, 2020, ISSN 0931-9085 ( taz.de [accessed on March 18, 2020]).
- ↑ Lars Fischer, Alina Schadwinkel: Does the coronavirus cause drug bottlenecks? Does the virus come from the pangolin? Website Spektrum.de , February 10, 2020, accessed on February 15, 2020 .
- ↑ Sixth infection confirmed: child in Bavaria fell ill with coronavirus. In: Website tagesschau.de . January 31, 2020, accessed February 9, 2020 .
- ↑ Donald Trump's test for coronavirus was negative. In: Der Spiegel - Politics. Retrieved March 15, 2020 .
- ↑ Florian Schumann: German coronavirus expert: "We have to prepare for a pandemic". In: Website Der Tagesspiegel . February 13, 2020, accessed February 15, 2020 .
- ↑ Coronavirus vaccine developers wary of errant antibodies www.nature.com, June 5, 2020
- ↑ Pathogenesis of oral type I feline infectious peritonitis virus (FIPV) infection: Antibody-dependent enhancement infection of cats with type I FIPV via the oral route www.ncbi.nlm.nih.gov, online publication April 23, 2019
- ↑ www.rki.de: Delivery bottlenecks .
- ↑ Health Senator welcomes expansion of the risk area through the Robert Koch Institute. In: berlin.de. February 26, 2020, accessed February 29, 2020 .
- ↑ Coronavirus: When does a disaster apply - and what follows? In: rbb24.de. February 26, 2020, accessed February 29, 2020 .
- ^ World Health Organization: Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). January 30, 2020, accessed on January 30, 2020 .
- ↑ Tedros Adhanom Ghebreyesus: WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020. In: WHO website . March 11, 2020, accessed on March 12, 2020 .
- ↑ Official justification in the draft law on BT-Drs. 19/18967 , p. 55.
- ↑ Protection package for more corona tests and care premiums decided. German Bundestag, May 14, 2020, accessed on May 14, 2020 .
- ↑ Non-namable reporting of examination results. Robert Koch Institute, June 4, 2020, accessed on June 8, 2020 .
- ^ Epidemic Act 1950. Consolidated Federal Law, Entire Legal Regulation. In: RIS . June 14, 2018, accessed on March 6, 2020 : "Subject to the reporting requirement: [...] (2) The Federal Minister for Health and Women can, [...], make other communicable diseases subject to the reporting requirement or expand existing reporting requirements."
- ↑ 15. Ordinance of the Federal Minister for Labor, Social Affairs, Health and Consumer Protection regarding notifiable communicable diseases 2020. In: Federal Law Gazette for the Republic of Austria. January 26, 2020, accessed on March 4, 2020 : "The obligation to notify according to the Epidemic Act 1950 is subject to suspected illnesses and deaths from 2019-nCoV (" 2019 novel coronavirus ")."
- ↑ Separation Ordinance. Consolidated federal law: Entire legal regulation for the segregation of sick people, suspected illnesses and contagion suspects and designation of houses and apartments, version dated March 6th, 2020. In: RIS, Federal Law Consolidated. January 31, 2020, accessed on March 6, 2020 : "In the case of measles or infection with 2019-nCoV (" 2019 novel coronavirus "), the sick and suspected diseases must be separated or, depending on the circumstances of the case, only subjected to certain traffic restrictions."
- ↑ Ordinance of January 31, 2020. 21. Ordinance of the Federal Minister for Social Affairs, Health, Care and Consumer Protection, with which the ordinance of the Minister of the Interior in agreement with the Minister of Education and Education of February 22, 1915, regarding the segregation of sick people, Disease suspect and contagion suspect and the names of houses and apartments is changed. In: Federal Law Gazette for the Republic of Austria. January 31, 2020, accessed on March 6, 2020 : "In § 4 3rd sentence, the word sequence" or infection with 2019-nCoV ("2019 novel coronavirus") "is inserted after the word" measles "."
- ↑ New Coronavirus: Information for Health Professionals. Suspicion and reporting criteria as well as reporting form. Federal Office of Public Health , March 5, 2020, accessed on March 5, 2020 .
- ↑ Federal Act on Combating Communicable Diseases in Humans. Epidemics Act, EpG. Federal Chancellery, January 1, 2017, accessed on March 6, 2020 .
- ^ Ordinance on combating communicable diseases in humans. (Epidemic Ordinance, EpV) of April 29, 2015 (as of March 1, 2019). Federal Chancellery, accessed on March 6, 2020 .
- ^ Ordinance of the FDHA on the reporting of observations of communicable diseases in humans. from December 1, 2015 (as of February 1, 2020). Federal Chancellery, accessed on March 6, 2020 .
- ↑ Registration forms. COVID-19 notification. Federal Office of Public Health, May 18, 2020, accessed on June 8, 2020 .